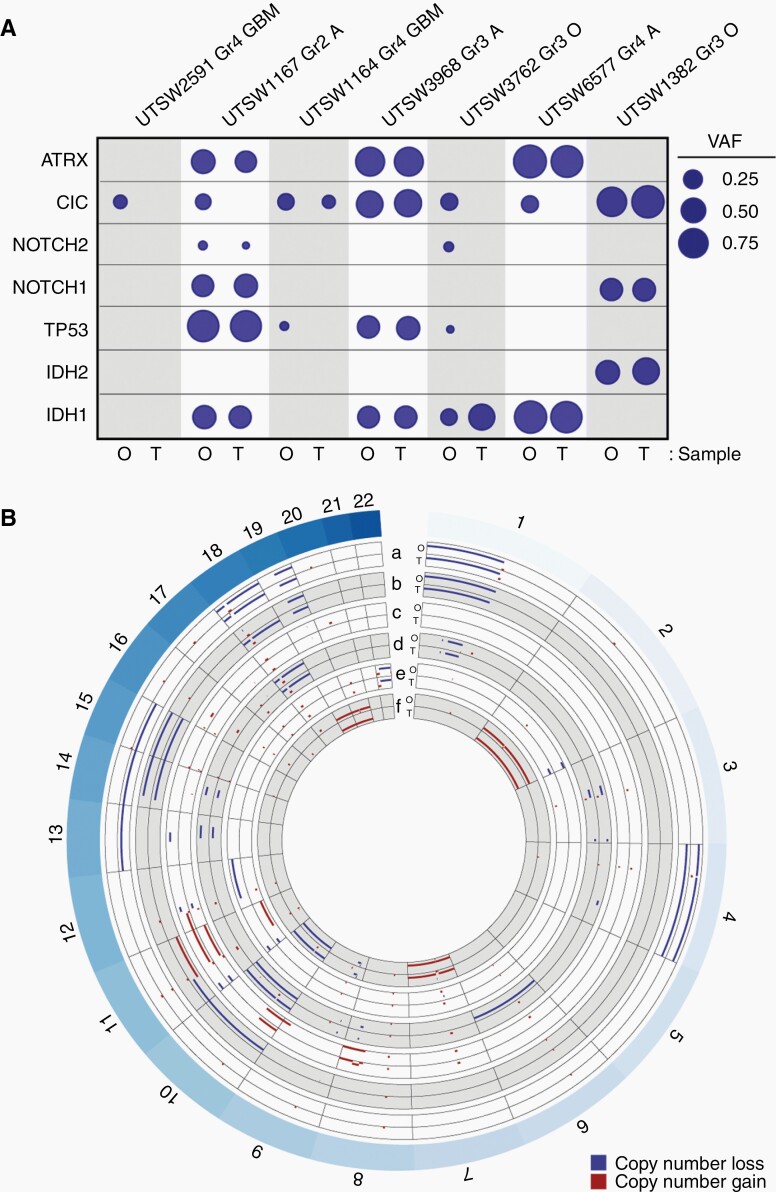
Fig. 5.
LGG organoids retain disease-specific genomic hallmarks. (A) LGG organoids largely retain key genetic mutations observed in their respective parental tumor specimens. Genomic DNA from organoid-primary tumor pairs was analyzed via targeted sequencing. Variant allele frequency (VAF) was calculated for each mutated gene. Mutations and single nucleotide variants that are not predicted to be pathogenic were excluded. Circle areas correspond to VAFs for each gene. Abbreviations at top of figure: Gr, Grade; GBM, glioblastoma; O, oligodendroglioma; A, astrocytoma. Abbreviations at bottom of figure: O: organoid, T: tumor. (B) LGG organoids retain copy number alterations observed in their respective parental tumor specimens. DNA sequencing data from part A was used to construct concentric Circos plots depicting copy number variations in paired organoid (O) and tumor (T) samples for the following tumors: a) oligodendroglioma, IDH-mutant, 1p/19q co-deleted, WHO Grade 3 (UTSW1382), b) oligodendroglioma, IDH-mutant, 1p/19q co-deleted, WHO Grade 3 (UTSW3762), c) astrocytoma, IDH-mutant, WHO Grade 3 (UTSW3968), d) GBM, IDH-wildtype, WHO Grade 4 (UTSW1164), e) astrocytoma, IDH-mutant, WHO Grade 2 (UTSW1167), f) GBM, IDH-wildtype, WHO Grade 4 (UTSW2591). Labels around circumference represent chromosome numbers. Scale of each axis ranges from copy number 0 (closer to exterior of the circle) to 4 (closer to interior of the circle), with any regions of copy number 2 not plotted. Red = gains, blue = losses. All samples with copy number >4 are plotted with copy number set to 4. Notably, 1p/19q co-deletion is observed in both organoid and primary tumor samples in a and b. Organoids were cultured for 4–6 weeks after explantation.