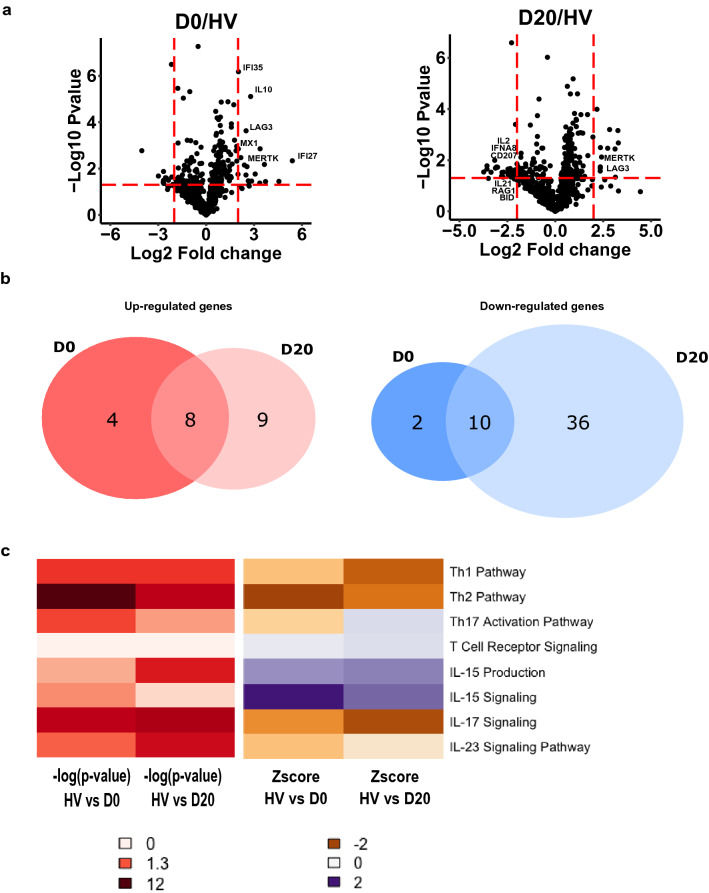
Fig. 1.
Transcriptomic profile of mononuclear cells in severe COVID-19 patients. RNA extracted from PBMCs of COVID-19 patients at day 0 and day 20 (n = 10) and healthy volunteers (HV, n = 10) was analyzed through NanoString technology. a Volcano plots of differentially expressed genes between patients sampled at D0 or at D20 and HV are showed. Limits of significance are illustrated by red dotted lines (i.e. Log2 Fold Change = −2 or + 2 and −Log 10 P value = 1.3). Selected genes are mentioned. b Venn diagrams of significantly up-regulated (left diagram, n = 21) or down-regulated (right diagram, n = 48) genes between patients and HV are showed. c Ingenuity Pathway Analysis was applied on the list of differentially expressed genes at D0 and D20. Heatmaps of Log 10 P-value (from white indicating the absence of significance to dark red indicating a strong significance) and Z-score (from orange indicating a down-regulation to purple indicating an up-regulation) for pathways related to T cell activation at D0 and D20 are presented