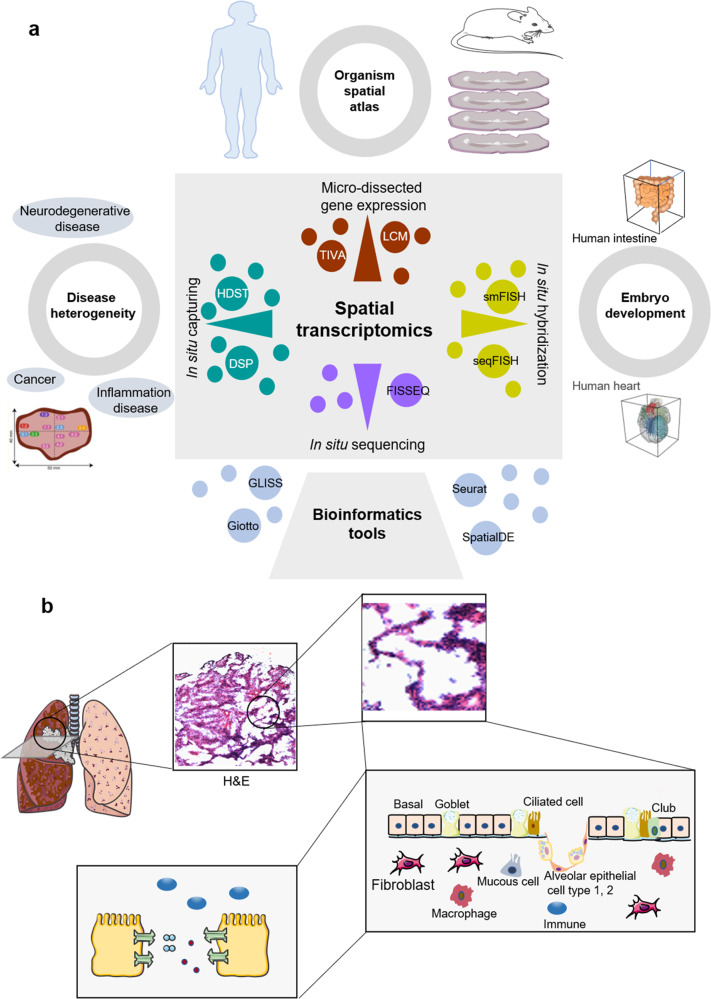
Fig. 6.
Summary of workflows during applications for spatial transcriptomics. a Spatial transcriptomics techniques are mainly used to solve spatial heterogeneity of diseases, biological spatial transcriptome map, and embryonic development spatial blueprint. The four ST techniques include micro-dissected gene expression, in situ hybridization, in situ sequencing and in situ capturing. Different techniques are used according to different sample characteristics and needs, and a variety of biological tools are used for analysis. b A simulation diagram of tumor microenvironment. ST technology assists in understanding the spatial location of tumor cells and gene expression as well as intercellular molecular communication between cells within the microenvironment. For example, H&E staining in lung tissue provides sample pathological information and assesses sample quality, and further integrates cell grouping and location information analysis such as basal, goblet, ciliated cell, alveolar epithelial cells type 1, 2 and other cells, so as to provide a better understanding of the molecular communication mechanisms in the cellular microenvironment