Figure 1. Identification of transcription factors required for pluripotency using the CRISPR TF library.
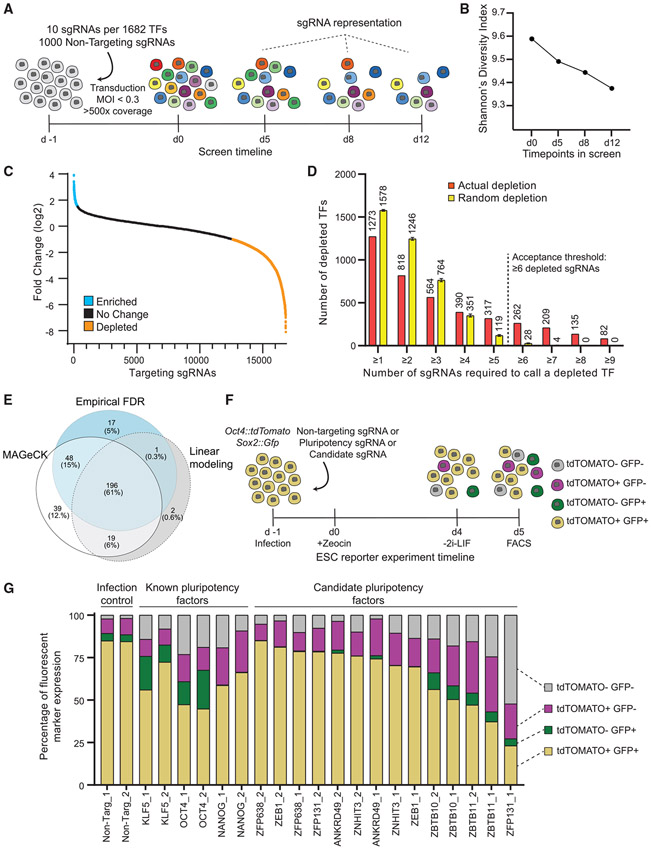
(A) Transcription factor screen overview. Day −1, CRISPR TF library transduction to growing ESCs; day 0, zeocin selection begins; days 5, 8, and 12, collection days and representation analysis.
(B) Shannon’s diversity index (H) was calculated based on the proportion of individual sgRNAs (pi) in the total number of sgRNAs in the pool (S) for each time point with sgRNA counts. The diversity score decreases as the proportion of essential and pluripotency-targeting sgRNAs decreases in the population.
(C) Log2 fold change of targeting sgRNAs on day 12 compared with the initial library representation. sgRNAs depleted more than the 50th most depleted non-targeting sgRNA are in orange and enriched more than the 50th most enriched non-targeting sgRNA are in blue.
(D) The number of targeting sgRNAs depleted and mean number of sgRNAs with random depletion calculated by 1,000 permutation tests for the different thresholds of sgRNA per gene. TFs with more than or equal to six depleted sgRNAs are unlikely to be false positives.
(E) Venn diagram representation of the number of depleted TFs identified by three different methods: log-fold change, linear modeling, and MAGeCK. All methods picked 196 genes as high-confidence depleted TFs.
(F) The workflow of the secondary screen using Oct4:tdTomato Sox2:Gfp pluripotency reporter ESC line. Pluripotent ESCs are depicted as yellow while differentiating cells lose one or both reporter expression.
(G) Percentage of cells expressing fluorescent reporters when transduced with a single sgRNA that is either non-targeting or targeting a known pluripotency TF or candidate TF. Both fluorescent reporters are expressed by 80% of cells transduced with non-targeting sgRNA. This number decreases in cells transduced with sgRNAs targeting known pluripotency TFs and Zbtb10, Zbtb11, and Zfp131.
See also Figures S1-S6 and Tables S1, S2, and S3.