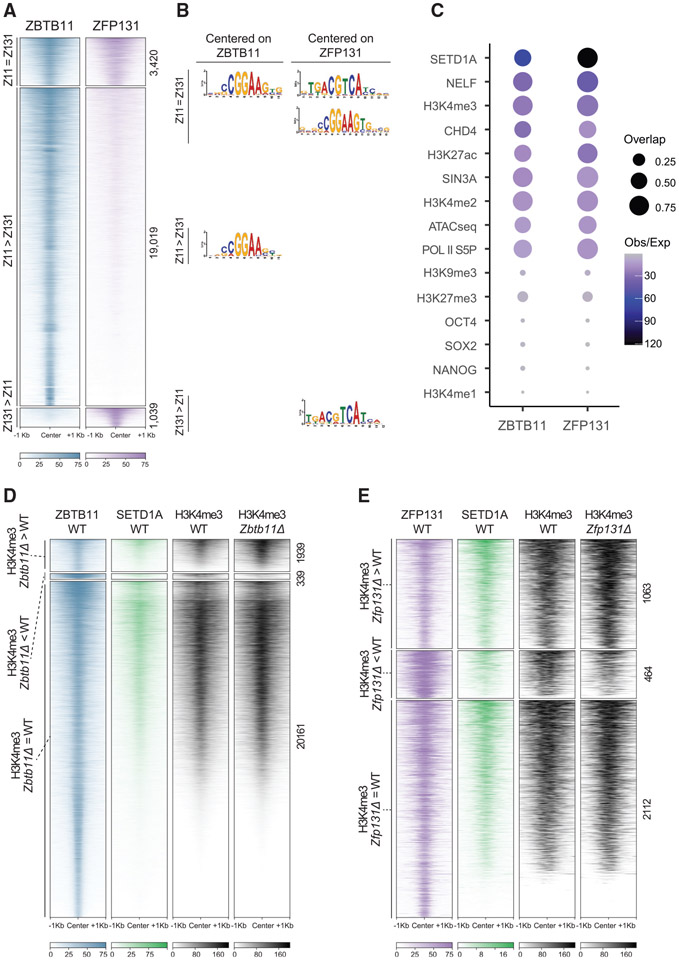
Figure 4. ZBTB11 and ZFP131 bound sites gain an H3K4me3 signal in Zbtb11Δ and Zfp131Δ knockout cells.
(A) ChIP-seq heatmap for ZBTB11 and ZFP131 binding on the genome ESC grown in +2i + LIF (n = 2). Categories were identified with ZBTB11 and ZFP131 co-binding (Z11 = Z131), differentially enriched ZBTB11 binding (Z11 > Z131), and differentially enriched ZFP131 binding (Z131 > Z11).
(B) Enriched motifs discovered via MEME-ChIP for each category of ZBTB11 and ZFP131 binding. Motif discovery was performed with centering to the peak at ZBTB11 and ZFP131 for three differential binding categories.
(C) Percentage of overlap between ZBTB11 and ZFP131 with pluripotency factors, known regulatory complex members, histone modifications, and ATAC-seq signal. ZBTB11 and ZFP131 bind at sites with active chromatin states, accessible and enriched with TrxG, Pol II S5P, NELF, and histone deacetylase members. ZBTB11 and ZFP131 do not co-bind with pluripotency TFs but bind to regions with active chromatin signatures. H3K4me1 data are obtained from GSE24165; H3K9me3 data are obtained from GSE18371; OCT4, NANOG, and SOX2 data are obtained from GSE11724; H3K4me2 data are obtained from GSE11172; and ATAC-seq data are obtained from GSE80511.
(D and E) ChIP-seq heatmap for (D) ZBTB11 and (E) ZFP131 overlap with SETD1A and H3K4me3. Categories are determined according to gain, loss, or no change in H3K4me3 signal between knockout and WT cells (n = 2 for all). ZBTB11 and ZFP131 co-occupy ESC genome with SETD1A. H3K4me3 comparison was done in −2i + LIF.
See also Figures S9 and S10.