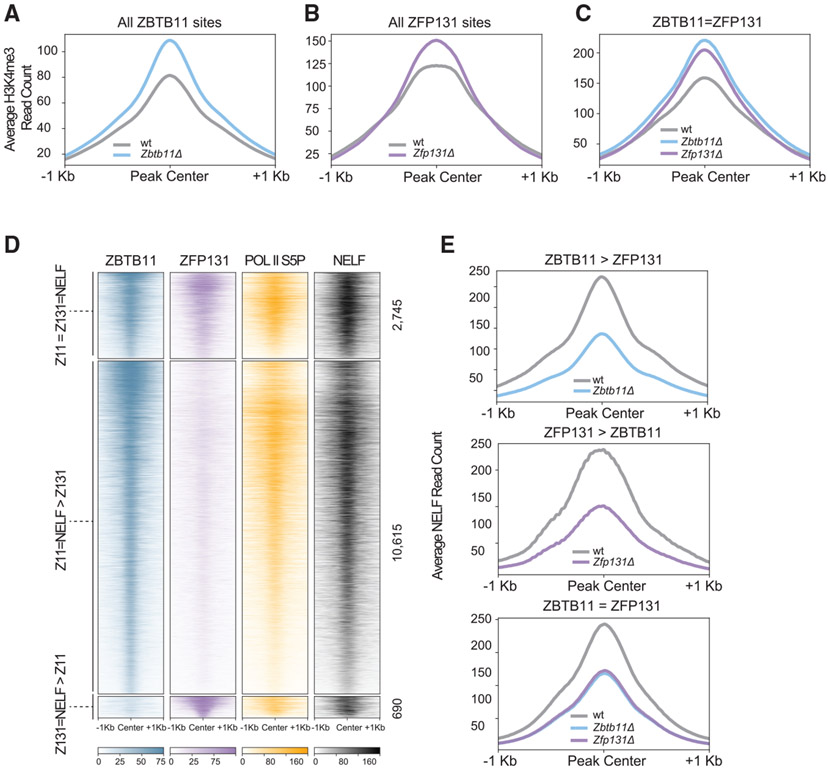
Figure 5. SETD1A occupancy at ZBTB11 and ZFP131 sites.
(A–C) Metagene plots for H3K4me3 difference (A) at ZBTB11-bound sites in Zbtb11Δ versus WT (n = 2), (B) at ZFP131-bound sites in Zfp131Δ versus WT (n = 2), and (C) at ZBTB11 and ZBTB131 co-bound sites in Zbtb11Δ and Zfp131Δ versus WT. Zbtb11 and Zfp131 mutations result in an H3K4me3 gain at ZBTB11 and ZFP131 binding sites. All comparisons were done in −2i + LIF.
(D) ChIP-seq heatmap for ZBTB11 and ZFP131 overlap with RNA Pol II S5P and NELF. Categories are determined according to ZBTB11 only, ZFP131 only, and co-bound regions (n = 2 for all). ZBTB11 and ZFP131 co-occupy ESC chromatin with RNA Pol II S5P and NELF.
(E) Metagene plots for NELF signal difference in ZBTB11 only, ZFP131 only, and co-bound regions (n = 2 for all). ZBTB11 and ZFP131 sites lose NELF in Zbtb11 and Zfp131 knockout ESCs. All comparisons were done in −2i + LIF.