Figure 3.
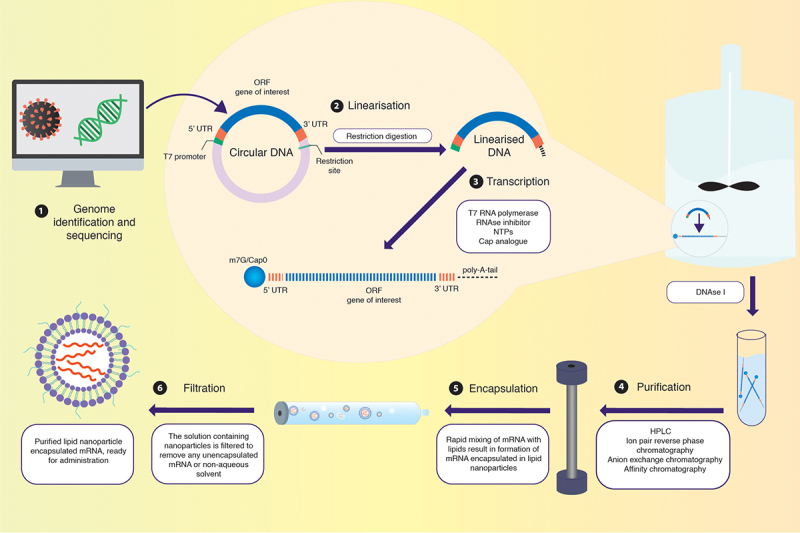
Synthesis of IVT-mRNA. (1) IVT-mRNA synthesis begins with a DNA template, usually pDNA, engineered to contain the gene of interest coding for the desired antigen identified by sequencing the genome of the target pathogen. (2) The pDNA template must include a bacteriophage promoter (T7), the ORF, a poly-deoxyribose T sequence (to code for the poly-A tail), and a restriction site which allows for restriction enzyme-mediated linearisation. (3) The linearised template undergoes in vitro transcription using a bacteriophage-derived T7 RNA polymerase. Following transcription, a mixture containing the desired mRNA, phage RNA polymerase, and nucleoside triphosphates is obtained. The 5’-cap can be introduced either enzymatically or be yielded in the transcription stage by including N7-methyl-guanosine analogue residues in excess of guanosine triphosphate (GTP) residues present. The 3’-tail can also be enzymatically added in this step, if not already incorporated. (4) The mixture is then purified using DNase I to degrade any contaminants and the template, followed by purification of the desired mRNA transcripts from a mix of abortive transcripts, longer transcripts with a 3’-overhang, oligodeoxynucleotides, and free nucleotides. While purification can be achieved by a series of precipitation and extraction steps, a chromatographic process is best suited for separating the transcripts of varied sizes. Techniques like high-performance liquid chromatography (HPLC) further refine the quality of the product, diminishing possibilities of unnecessarily activating innate immune sensors via contaminants. (5) Rapid mixing of the obtained mRNA with lipid via the utilisation of microfluidics allows for the self-assembly of mRNAs within lipid nanoparticles. (6) The obtained solution containing nanoparticles has to undergo further dialysis or filtration to remove any unencapsulated mRNA or non-aqueous solvent. Post filtration, the purified solution containing mRNA within lipid nanoparticles is ready for administration within the host.
