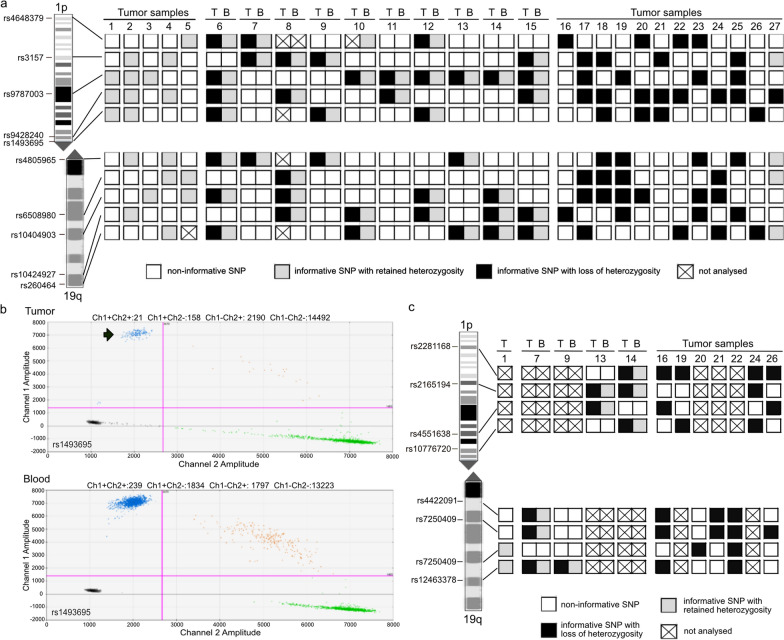
Fig. 2.
Loss of heterozygosity (LOH) on chromosomal arms 1p and 19q detected by ddPCR-based SNP analysis. a Patterns of ddPCR-based SNP analyses for LOH on 1p and 19q in 27 gliomas, previously shown to either have retained (tumor samples 1–5 and 27) or lost 1p and 19q (tumor samples 6–26). In ten patients with IDH-mutant and 1p/19q-codeleted oligodendrogliomas, matched DNA samples extracted from tumor tissue (T) and blood leucocytes (B) were investigated. In the remaining 17 patients, only T DNA samples were investigated. The cases 1–5 without 1p/19q codeletion were used to establish the cut-off value for the detection of LOH. Note that all patients showed one or more informative (heterozygous) SNPs on 1p, while 3 of the 27 patients lacked an informative SNP on 19q. Presence or absence of 1p/19q-codeletion in each individual case was validated by an independent method. White rectangles, non-informative SNP; light grey rectangles, informative SNP with retained heterozygosity; black rectangles, informative SNP with loss of heterozygosity; crossed rectangles, data not evaluable. b Exemplary presentation of a two-dimensional plot generated by the Quantasoft™ Software (Bio-Rad Laboratories). Shown are the results of the ddPCR-based analysis of SNP rs1493695 in case 9, who exhibited a LOH at this locus. X axis, fluorescence intensity detected in the HEX-channel (channel 2); Y axis, fluorescence intensity detected in the FAM-channel (channel 1); pink line, threshold; grey dots, droplets with background fluorescence; green dots, droplets with fluorescence detected in the HEX-channel; blue dots, droplets with fluorescence detected in the FAM-channel; orange dots, droplets with signals in both channels. The tumor-DNA showed a reduced number of droplets in the FAM-channel (arrow) compared to the number of droplets in the HEX-channel (green dots). The blood-derived DNA of this patient is heterozygous for this SNP with nearly the same number of droplets counted in the FAM- and HEX-channel. c Results of ddPCR analyses of additional SNPs located on 1p or on 19q in 12 selected cases, including 10 tumors with only one informative locus on one or both chromosome arms and two of the cases that were not informative at the five initially studied SNPs on 19q (see Fig. a above). Note that all cases demonstrated additional informative SNPs that confirmed the initial results and allowed for complete 1ß/19q copy number evaluation in the two cases that initially were not informative on 19q