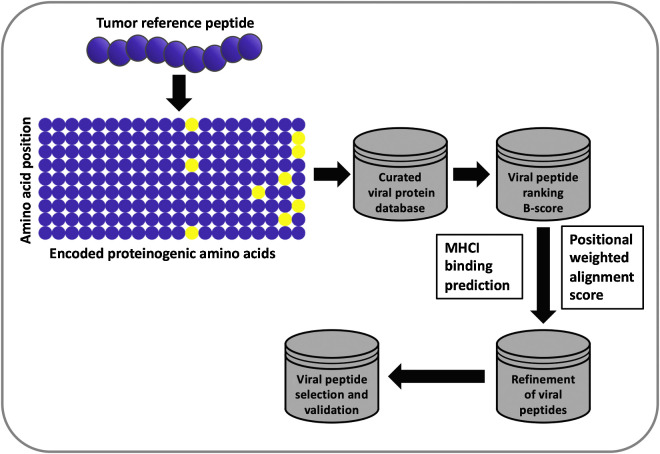
Figure 1.
Flowchart of the HEX algorithm. When using the HEX software, a matrix is generated on the basis of the amino acid composition of a tumor peptide (referred to as the reference peptide). This matrix is then used to scan the viral database, and resulting viral peptides are ranked in order of log-likelihood of recognition (B-score). Each viral peptide is assigned an alignment score and a score for the predicted MHC class I–binding affinity. The candidate viral peptides are ranked on the basis of the following criteria: MHC class I–binding affinity prediction score > alignment score > B-score; the highest scoring peptides are analyzed experimentally.