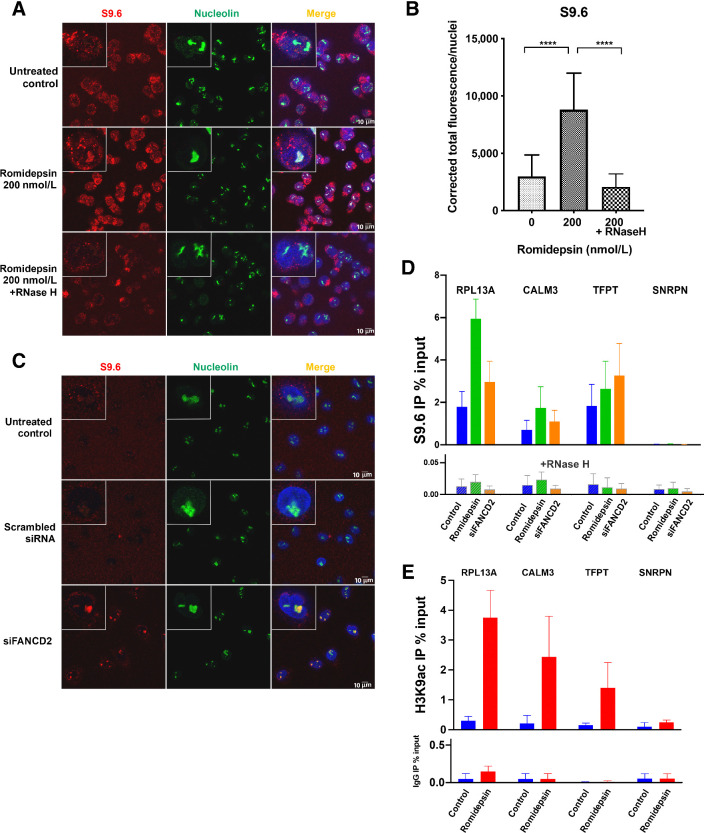
Figure 4.
R-loop structures form following romidepsin treatment. A, DNA–RNA hybrid accumulation in romidepsin-treated cells. Staining was carried out using S9.6 and nucleolin antibodies. Nuclei were stained with DAPI. Bottom, treatment with RNase H in the romidepsin-treated cells eliminated the DNA–RNA hybrids. B, Quantification of nuclear S9.6 immunofluorescence signal in LOXIMVI cells treated with or without romidepsin and with RNase H following romidepsin treatment. The S9.6 signal was quantified only in the nuclear regions, DAPI staining. The median of the nuclear S9.6 signal intensity per nucleus is shown. ***, P < 0.001; ****, P < 0.000 (t test, two‐tailed). C, Detection of DNA–RNA hybrids in siFANCD2-transfected A549 cells. Top two demonstrating the untreated control and negative control of siRNA experiment and the bottom is showing positive R-loop staining in siFANCD2 cells. See also Supplementary Fig. S4A. D, DRIP–qPCR signal values at RPL13A, CALM3, TFPT, and SNRPN genes in LOXIMVI cells treated with or without romidepsin for 6 hours. LOXIMVI cells transfected with the FANCD2 siRNAs were included as positive control for R-loops. Cells treated in vitro with (bottom) or without (top) RNase H before immunoprecipitation. The mean ± SEM from three independent experiments is shown. E, ChIP-qPCR was performed with antibody against H3K9ac in the LOXIMVI cells after treatment with romidepsin for 6 hours. Immunoprecipitated chromatin samples were analyzed by qPCR using specific primer pairs as shown on Supplementary Table S1. The mean ± SEM from three independent experiments is shown.