Abstract
OBJECTIVE.
The purpose of this study was to evaluate in a multicenter dataset the performance of an artificial intelligence (AI) detection system with attention mapping compared with multiparametric MRI (mpMRI) interpretation in the detection of prostate cancer.
MATERIALS AND METHODS.
MRI examinations from five institutions were included in this study and were evaluated by nine readers. In the first round, readers evaluated mpMRI studies using the Prostate Imaging Reporting and Data System version 2. After 4 weeks, images were again presented to readers along with the AI-based detection system output. Readers accepted or rejected lesions within four AI-generated attention map boxes. Additional lesions outside of boxes were excluded from detection and categorization. The performances of readers using the mpMRI-only and AI-assisted approaches were compared.
RESULTS.
The study population included 152 case patients and 84 control patients with 274 pathologically proven cancer lesions. The lesion-based AUC was 74.9% for MRI and 77.5% for AI with no significant difference (p = 0.095). The sensitivity for overall detection of cancer lesions was higher for AI than for mpMRI but did not reach statistical significance (57.4% vs 53.6%, p = 0.073). However, for transition zone lesions, sensitivity was higher for AI than for MRI (61.8% vs 50.8%, p = 0.001). Reading time was longer for AI than for MRI (4.66 vs 4.03 minutes, p < 0.001). There was moderate interreader agreement for AI and MRI with no significant difference (58.7% vs 58.5%, p = 0.966).
CONCLUSION.
Overall sensitivity was only minimally improved by use of the AI system. Significant improvement was achieved, however, in the detection of transition zone lesions with use of the AI system at the cost of a mean of 40 seconds of additional reading time.
Keywords: artificial intelligence, laparoscopic, MRI, multiparametric, prostate cancer, radical prostatectomy, robot-assisted
Prostate cancer is the most common noncutaneous cancer type among men [1]. Unlike most other cancers, prostate cancer is difficult to detect with conventional imaging techniques such as ultrasound and CT. Therefore, until recently, imaging has not been accepted as standard-of-care practice for prostate cancer detection. Over the last 2 decades, major advances in prostate MRI have led to considerable improvements in prostate cancer detection. Although initially the use of MRI was limited [2], with the development of higher magnetic field strengths, higher quality of imaging, and the combined use of anatomic and functional MRI sequences, multiparametric MRI (mpMRI) has emerged as an important method of detecting prostate cancer [3]. Reports in the current literature, however, indicate that 5–30% of prostate cancers are missed at mpMRI [4–6]. The causes of such misses may be related to the complex nature of the prostate tissues and the limited spatial resolution of MRI. One proposed solution is to use artificial intelligence (AI)-based detection systems.
With the help of machine learning, classification algorithms can be trained to predict results and outcomes, provided that enough training data are available. In 2017, we at the National Cancer Institute [7] proposed an AI system based on intensity and texture analysis and a random forest classification algorithm. This system was validated in a large multireader multicenter study in 2018 [8]. Results of that study revealed an increase in detection of transition zone lesions among moderately experienced readers only. Overall, however, the AI system was equivalent to conventional MRI interpretation [8]. In that study, color-coded prediction maps were used to draw attention to AI-detected lesions. Feedback from the study suggested that prediction maps compromised the interaction between the radiologists and the AI system with resultant decreased accuracy for some readers. To address this issue a new AI detection system with more expert annotated data was designed. Instead of color-coded cancer probability maps as output, the new AI system provides attention map boxes encompassing areas of increased likelihood of prostate cancer. Herein we report the results of our multicenter multireader study of the AI system with this new user interface. The main objective of the study was to evaluate in a multicenter dataset the performance of an AI detection system with attention map boxes compared with mpMRI interpretation for detection of prostate cancer.
Materials and Methods
Patient Population
This HIPAA-compliant evaluation of multi-institutional data was approved by the National Cancer Institute ethics committee. Inclusion of anonymized data from the other institutions was approved in concordance with National Institutes of Health Office of Human Subjects Resources protocol 11617. Local ethics approvals to share data were obtained as needed. Patients from five institutions were included in this study. Those in the case group underwent mpMRI and had subsequent prostate biopsy results positive for adenocarcinoma and then underwent radical prostatectomy. Final histopathologic results for the prostatectomy specimens with lesion mapping were available for all case population patients. All participants in the control group underwent mpMRI with no visible lesions detected. Prostate cancer was ruled out by means of 12-core transrectal systematic biopsy. Three patients with missing final radical prostatectomy histopathologic lesion maps were excluded. The final study population included 152 case and 84 control subjects. The distribution of case and control subjects among institutions is summarized in Table 1.
TABLE 1:
Distribution of Patients and Control Subjects by Participating Institution
| Institution No. | Case Subjects | Control Subjects | Total |
|---|---|---|---|
| 1 | 22 | 14 | 36 |
| 2 | 24 | 17 | 41 |
| 3 | 30 | 21 | 51 |
| 4 | 31 | 23 | 54 |
| 5 | 45 | 9 | 54 |
|
| |||
| Total | 152 | 84 | 236 |
Reader Profiles
To prevent bias, the case and control MRI data were obtained from five different institutions but were interpreted by nine readers from independent institutions. Readers ranged in experience in interpreting prostate mpMRI and were stratified into three levels of experience: low, moderate, and high. Determination was based on years of experience and number of prostate MRI studies read per year according to the following criteria: a low level of experience was less than 1 year or fewer than 100 examinations per year; moderate, 1–3 years or 100–300 examinations per year; high, more than 3 years or more than 300 examinations per year.
Study Design and Statistical Powering
Because it was unrealistic for every reader to read every case, each patient imaging examination was randomly assigned to three different readers to ensure balanced and unbiased distribution. Our primary hypothesis was that AI-assisted mpMRI can achieve higher lesion-based sensitivity than mpMRI without the benefit of AI. To test this hypothesis, nine readers read assigned cases according to the balanced incomplete block design, in which each randomly selected case stratified by patient disease status was assigned to each triple-wise combination of readers [9]. Among the 236 patients (152 with cancer, 84 without cancer), each reader evaluated a mean of 78 patients (range, 75–81). The primary endpoint was the difference in mean lesion-level reader-specific sensitivity between AI-assisted detection and mpMRI alone. In the power analysis, sensitivity was set at 57% and the SD of the primary endpoint was estimated from previous studies [8, 10]. This study had 93% power to detect an 8% difference in sensitivity using the Z test at the two-sided 5% significance level.
MRI Acquisition Technique
All imaging examinations were performed at 3 T without an endorectal coil with equipment from a variety of vendors. Axial T2-weighted imaging, DWI with at least two b values, and dynamic contrast-enhanced imaging sequences were performed. Because DWI with a b value of 1500 mm/s2 was necessary for the AI processing, in cases in which this acquisition was not available, the high b value was calculated by use of a mono-exponential decay model. Image acquisition protocols were compliant with Prostate Imaging Reporting and Data System version 2 (PI-RADSv2) technical recommendations.
Patient Data Deidentification
In compliance with U.S. Office of Human Subjects Resources guidelines, all medical images were fully anonymized by provider centers before submission to our center (National Cancer Institute). For this purpose, standard scripts were used that remove all DICOM tags except for technical image acquisition–related information. When the data were received, a second deidentification was performed to ensure patient confidentiality at the highest standard.
Artificial Intelligence System
The AI system was based on a custom multitask random forest similar to the Hough forest and the regression forest [11]. Each random tree was trained on 3-T MR images of 161 patients from five different institutions; an alternating information gain function was used that was either defined to optimize for classification accuracy or minimize the l2 residual of predicted bounding box extents. The training population was patients and institutions different from those of the test population in this study. This learning system entailed a combination of patch-based intensity and Haralick texture features and operated only on presegmented transition and peripheral zones.
Automatic segmentations were performed on T2-weighted images and manually corrected by an expert radiologist. Each zone had its own specialized AI model. The result was a collection of 10 random trees per zone that each evaluated an image patch from T2-weighted images, apparent diffusion coefficient maps, and b1500 images and predicted both the probability of clinically significant cancer and the bounding box width and height of the lesion. The predictions from all trees were averaged, and box candidates were postprocessed with nonmaximum suppression to choose the final predicted boxes (up to four were kept). The result was a probability map for clinically significant cancer (Gleason score > 3 + 3) and a collection of attention map boxes for suspicious lesions, the latter of which were used in this study. The attention map boxes were picked by use of a threshold corresponding to a 67% tumor detection rate at 2.71 false-positive results per patient.
A pixel-based cancer probability map was calculated, and for this study, the readers were provided with a maximum of four attention map boxes corresponding to regions of high cancer probability, which were overlaid on the T2-weighted image from each case and control MRI examination. The idea behind this approach was to ensure that readers can uniformly focus on the most suspicious possible lesions on MRI studies without interfering with the actual image, as occurs with conventional probability maps, which usually cover the underlying image. These maps were termed attention maps to distinguish them from probability maps (Fig. 1).
Fig. 1—
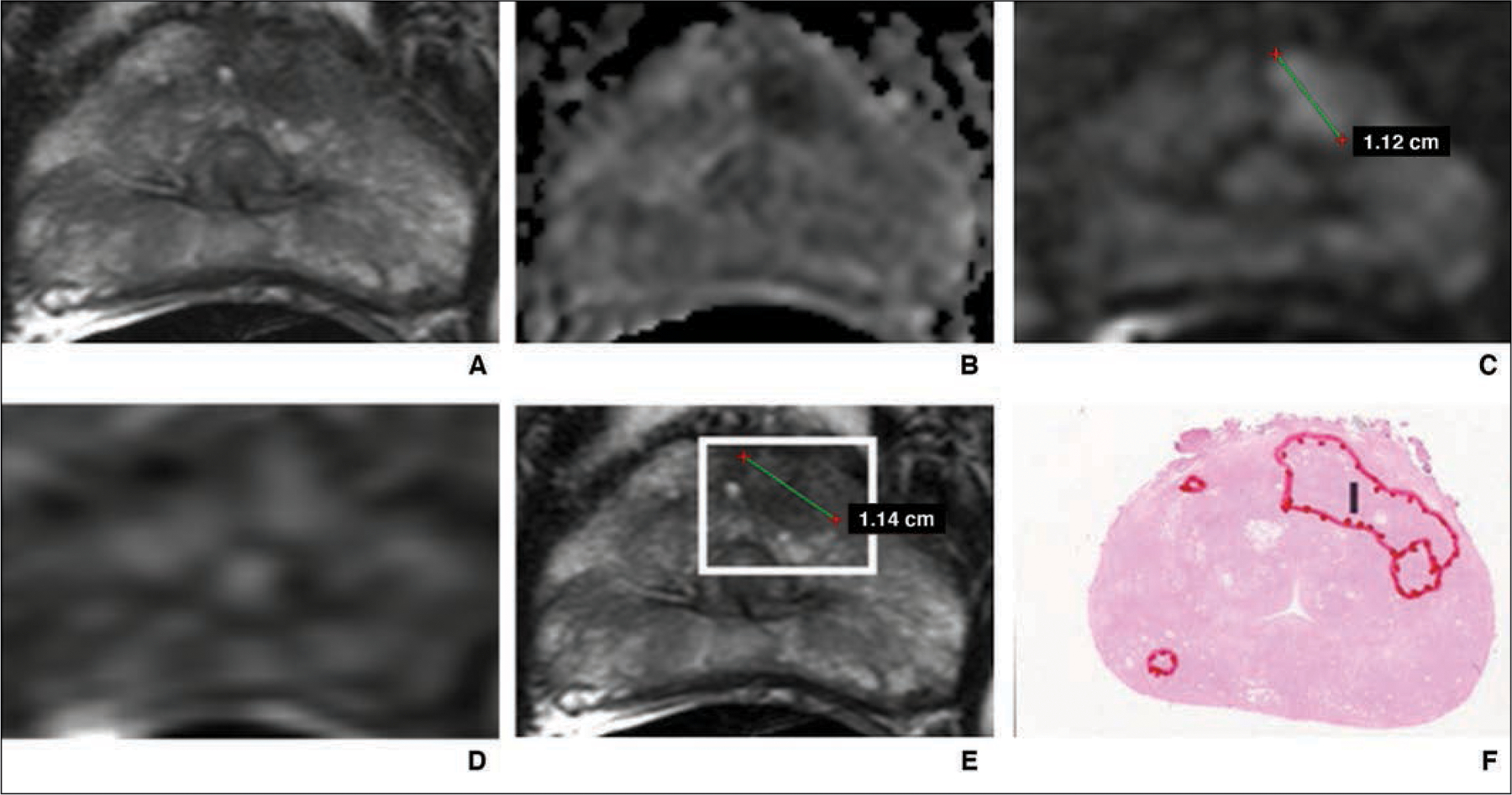
55-year-old man with prostate-specific antigen level of 4.68 ng/mL and Prostate Imaging Reporting and Data System category 5 lesion in left anterior transition zone correctly detected by artificial intelligence system. Final histopathologic result was Gleason 3 + 4 prostate cancer.
A, T2-weighted MR image.
B, Apparent diffusion coefficient map.
C, DW image (b = 2000 mm/s2).
D, Dynamic contrast-enhanced MR image.
E, T2-weighted MR image with attention box produced by means of artificial intelligence.
F, Photomicrograph of radical prostatectomy specimen. I = index lesion.
Image Evaluation
All readers used a commercially available DICOM viewer (RadiAnt, Medixant) at their personal workstations. Readers were blinded to clinical and histopathologic outcomes.
In the first session, T2-weighted, DWI (apparent diffusion coefficient, b = 1500 mm/s2), and dynamic contrast-enhanced images were presented to the readers for tumor detection and evaluation. For each patient a database application (Access, Microsoft Office 365) readout form with a pseudoidentifier was provided for documentation and analysis. Readers could call up to four lesions per image and assign a PI-RADSv2 category for each detected lesion. In addition, the location of each lesion was documented in accordance with the PI-RADSv2 recommendations, and a screen shot of the lesion was stored in the database document [12]. A timer recorded and saved the reading time for each reader per study.
After a 4-week washout period, a training package was sent to participants with three examples, so that they could become familiar with handling and interpretation of the AI system. Readers were instructed to read the AI images first and assess each attention box. Thereafter, the location of the boxes was annotated on the corresponding mpMR images. During the AI-assisted readout session, the participants accepted a lesion if its PI-RADSv2 category within the attention box was 3 or greater or rejected it if the PI-RADSv2 category was less than 3. The readers were prohibited from evaluating lesions detected on mpMR images other than those in attention boxes provided by the AI system. This stringent approach, defined as first-reader design workflow, would theoretically simulate the raw performance of AI as used by radiologists [13]. The results of the reading session were stored in a database readout form similar to that used in session 1.
Histopathologic Assessment
The genitourinary pathologist at each provider center was blinded to the mpMRI results. For each case patient’s specimen, cancer lesions were mapped, and a corresponding Gleason score according to the International Society of Urological Pathology 2014 consensus guidelines was assigned [14].
Statistical Analysis
Patient-based sensitivity and specificity were calculated at each PI-RADSv2 threshold, and AUC was estimated for each reader; the maximum PI-RADSv2 category assigned by a given reader represented each patient’s outcome. The comparison between AI and mpMRI was made at PI-RADSv2 1 or greater representing all detected lesions and PI-RADSv2 3 or greater representing suspicious lesions. For lesion-based analysis, reader sensitivity and free-response ROC analysis was performed [15]. Reader statistics were averaged across all readers and by experience level. Reader agreement on lesion detection in the same location was assessed by the index of specific agreement [16]. Statistical inference was obtained by the bootstrap resampling procedure with 2000 bootstrap samples drawn at the patient level. The 95% confidence limits were the 2.5% and 97.5% percentiles of the bootstrap resampling distribution. All test statistics were based on two-sided Wald test and bootstrap standard error. Values of p < 0.05 were considered statistically significant.
Results
Study Population and Lesion Characteristics
The final study population consisted of 152 case and 84 control subjects. Except in 38 case patients, the final histopathologic result was grade group 2 or higher. There were 274 pathologically proven cancer lesions with 188 of the 274 in the peripheral zone, 77 in the transition zone, and nine spanning both zones. Among all 274 lesions, 38 were assigned grade group 1, 130 group 2, 45 group 4, and 15 group 5 at final histopathologic analysis.
Multiparametric MRI and Artificial Intelligence Performance at Patient Level
The overall AUCs were 81.6% for MRI and 78% for AI (p = 0.053). Readers with a low experience level had AUCs of 80.9% for MRI and 73.3% for AI (p = 0.018); moderate experience, 80% for MRI and 76.9% for AI (p = 0.28); and a high level of experience, 83.8% for MRI and 83.6% for AI (p = 0.95).
Sensitivity and specificity plotted against PI-RADSv2 thresholds are shown in Figure 2. For the detection of all ground truth lesions (threshold, PI-RADSv2 ≥ 1), no significant difference in sensitivity was observed between MRI and AI (89.6% vs 87.9%, p = 0.364). However, in the subgroup of experienced readers, sensitivity of AI was significantly greater than that of MRI (95.5% vs 89.0%, p = 0.013). For lesions considered suspicious with MRI (threshold, PI-RADSv2 ≥ 3) no significant differences in sensitivity were observed in the whole group (81.7% vs 83.5%, p = 0.453). This finding held true for all subgroups of reader experience.
Fig. 2—
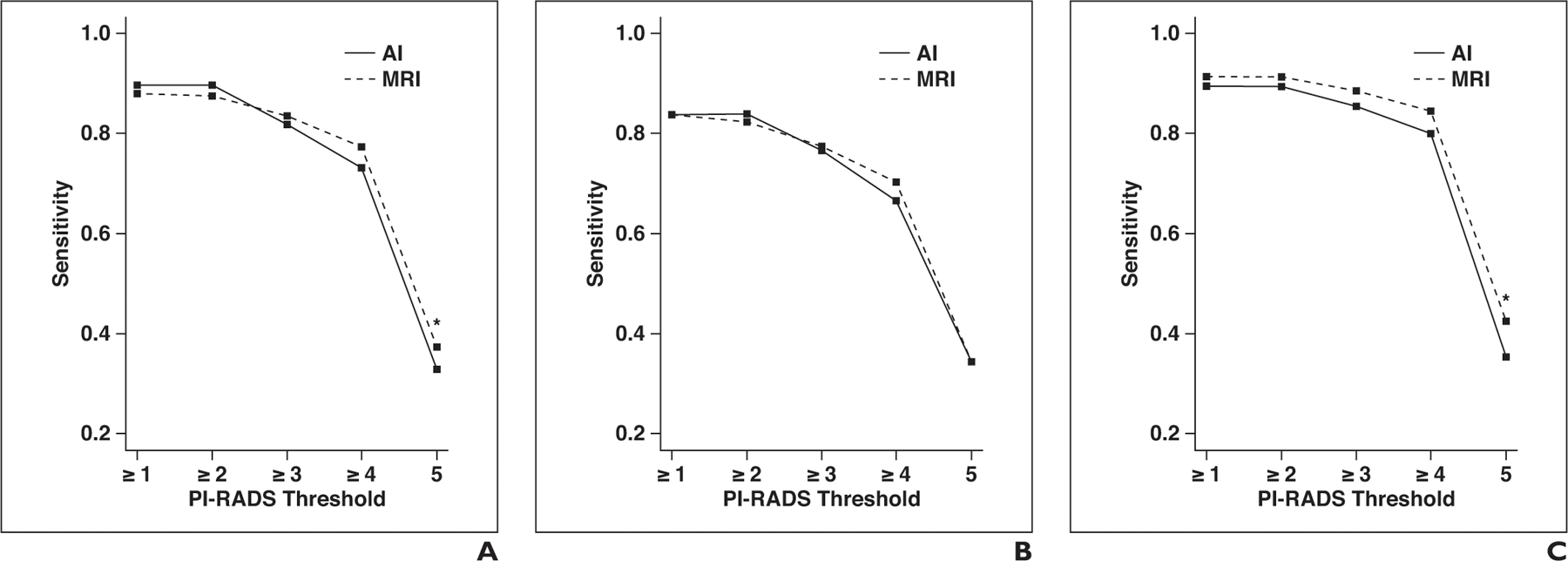
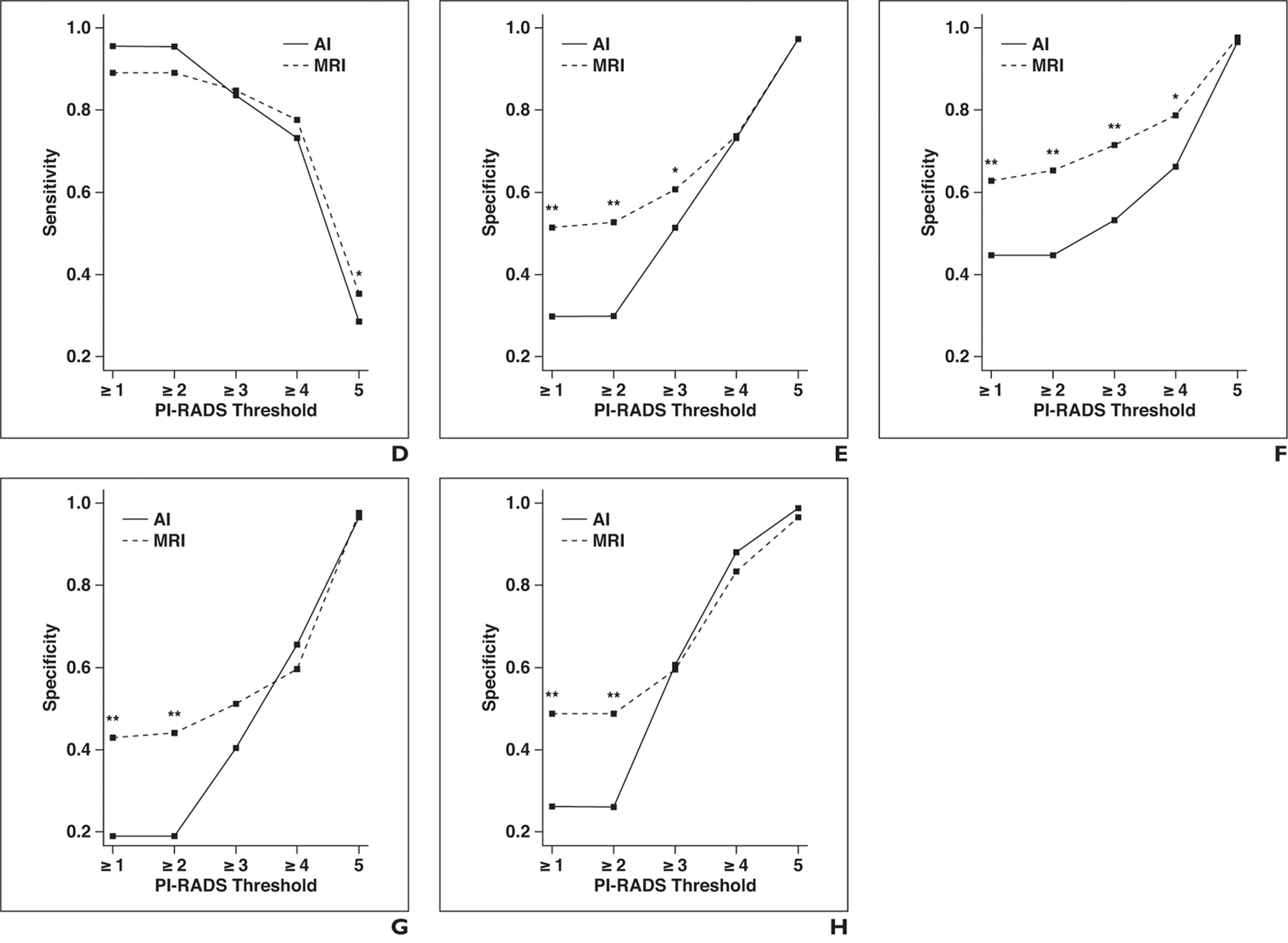
Graphs show sensitivity and specificity of artificial intelligence (AI) and MRI for different Prostate Imaging Reporting and Data System (PI-RADS) category thresholds at patient level. Asterisk denotes p < 0.05; double asterisk, p < 0.01.
A, Sensitivity for all readers.
B, Sensitivity for readers with low level of experience.
C, Sensitivity for readers with moderate level of experience.
D, Sensitivity for readers with high level of experience.
E, Specificity for all readers.
F, Specificity for readers with low level of experience. G, Specificity for readers with moderate level of experience.
H, Specificity for readers with high level of experience.
For the detection of all ground truth lesions (threshold, PI-RADSv2 ≥ 1), specificity was significantly lower for AI (30.0% vs 51.5%, p < 0.001) in the whole group. This finding held true for all subgroups of reader experience. For lesions considered suspicious on MRI studies (threshold, PI-RADSv2 ≥ 3), specificity was significantly lower for AI in the whole group (51.4% vs 60.7%, p = 0.01). Although it was observed in all subgroups of reader experience, this result did not reach statistical significance in the groups of readers with moderate and high experience levels.
Multiparametric MRI and Artificial Intelligence Performance at Lesion Level
The free-response ROC AUCs were 74.9% for MRI and 77.5% for AI (p = 0.095). Readers with a low experience level had AUCs of 76.6% for MRI and 78.4% for AI (p = 0.095); moderate experience, 78.1% for MRI and 78.6% for AI (p = 0.747); and a high experience level, 76.9% for MRI and 81.1% for AI (p = 0.003).
Lesion-level sensitivity and positive predictive value for all PI-RADSv2 thresholds are shown in Table 2 for MRI and AI. Sensitivity plotted against PI-RADSv2 thresholds for the whole prostate, peripheral zone, and transition zone are shown in Figure 3. For the detection of all ground truth lesions (threshold, PI-RADSv2 ≥ 1), sensitivity was higher for AI than for MRI but did not reach a statistical significance (57.4% vs 53.6%, p = 0.073). However, in the subgroup of highly experienced readers, AI had significantly greater sensitivity than MRI did (62.7% vs 54.1%, p = 0.002). There were no statistically significant differences between AI and MRI in the subgroups of readers with low and intermediate experience levels. For lesions considered suspicious on MRI studies (threshold, PI-RADSv2 ≥ 3), there was no significant difference in sensitivity between AI and MRI (50% vs 51%, p = 0.65). This was also true among all subgroups of reader experience.
TABLE 2:
Lesion-Level Diagnostic Performance Statistics on Artificial Intelligence (AI) and MRI for PI-RADS Category Thresholds
| PI-RADS Threshold for Performance Metric Evaluation | AI |
MRI |
||||||
|---|---|---|---|---|---|---|---|---|
| Overall | Reader Experience Level |
Overall | Reader Experience Level |
|||||
| Low | Moderate | High | Low | Moderate | High | |||
| PI-RADS ≥ 1 | ||||||||
| Sensitivity (%) | 57.4 (52.6–63.0) | 51.5 (44.4–59.1) | 58.0 (50.8–65.6) | 62.7 (56.9–69.9)a | 53.6 (48.5–59.6) | 49.9 (43.4–57.2) | 56.7 (50.3–64.2) | 54.1 (47.5–62.6)a |
| PPV (%) | 46.6 (42.9–50.4) | 51.9 (46.0–58.4) | 39.6 (33.9–45.1) | 48.2 (42.9–53.7) | 60.7 (56.9–64.6) | 62.1 (55.7–68.9) | 54.7 (48.8–60.7) | 65.2 (58.9–71.5) |
| PI-RADS ≥ 2 | ||||||||
| Sensitivity (%) | 57.4 (52.6–63.0) | 51.5 (44.4–59.1) | 58.0 (50.8–65.6) | 62.7 (56.9–69.9)a | 53.6 (48.5–59.6) | 49.9 (43.4–57.2) | 56.7 (50.3–64.2) | 54.1 (47.5–62.6)a |
| PPV (%) | 46.7 (43.0–50.7) | 51.9 (46.0–58.4) | 39.8 (34.1–45.3) | 48.5 (43.3–54.1) | 61.3 (57.4–65.3) | 63.7 (57.3–70.6) | 55.1 (49.4–61.0) | 65.2 (58.9–71.5) |
| PI-RADS ≥ 3 | ||||||||
| Sensitivity (%) | 50.0 (44.6–56.4) | 45.4 (39.1–53.0) | 52.6 (45.4–60.8) | 51.9 (44.5–60.5) | 51.0 (45.8–56.9) | 47.8 (41.1–55.3) | 54.0 (47.1–61.8) | 51.0 (44.3–59.3) |
| PPV (%) | 57.6 (53.6–61.7) | 56.3 (49.8–63.4) | 51.0 (45.2–57.0) | 65.6 (59.6–71.4) | 65.7 (61.7–69.7) | 68.2 (61.5–75.1) | 56.8 (50.7–63.2) | 72.0 (66.1–77.6) |
| PI-RADS ≥ 4 | ||||||||
| Sensitivity (%) | 42.3 (37.4–48.7) | 38.9 (32.9–46.3) | 45.2 (38.5–53.5) | 42.9 (36.5–50.3) | 45.1 (39.7–51.2) | 41.8 (35.2–49.8) | 49.4 (42.5–57.5) | 44.0 (37.1–52.2) |
| PPV (%) | 68.9 (64.7–73.1) | 62.4 (54.8–70.5) | 64.0 (57.3–71.0) | 80.2 (74.6–85.8) | 71.4 (67.4–75.8) | 71.0 (64.3–78.3) | 60.6 (54.2–67.2) | 82.6 (76.5–88.4) |
| PI-RADS ≥ 5 | ||||||||
| Sensitivity (%) | 18.3 (14.6–22.9) | 21.4 (16.3–27.4) | 18.3 (13.0–24.7) | 15.3 (10.7–21.1) | 19.2 (15.0–24.0) | 18.3 (13.1–24.4) | 21.4 (15.6–28.2) | 17.9 (13.0–24.4) |
| PPV (%) | 88.3 (83.2–93.3) | 89.8 (81.8–97.4) | 83.6 (74.6–91.9) | 91.5 (83.7–98.3) | 86.6 (81.3–92.0) | 86.1 (77.8–94.0) | 84.5 (74.7–94.3) | 89.3 (83.3–95.4) |
Note—Values in parentheses are 95% CI. PI-RADS = Prostate Imaging Reporting and Data System, PPV = positive predictive value.
p < 0.01.
Fig. 3—
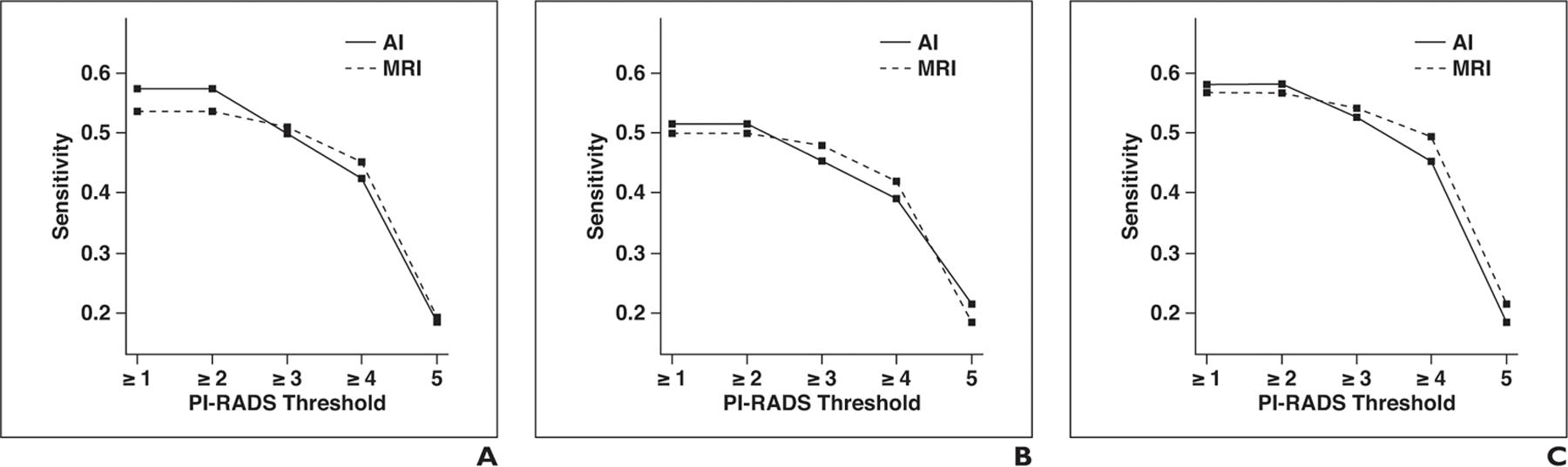
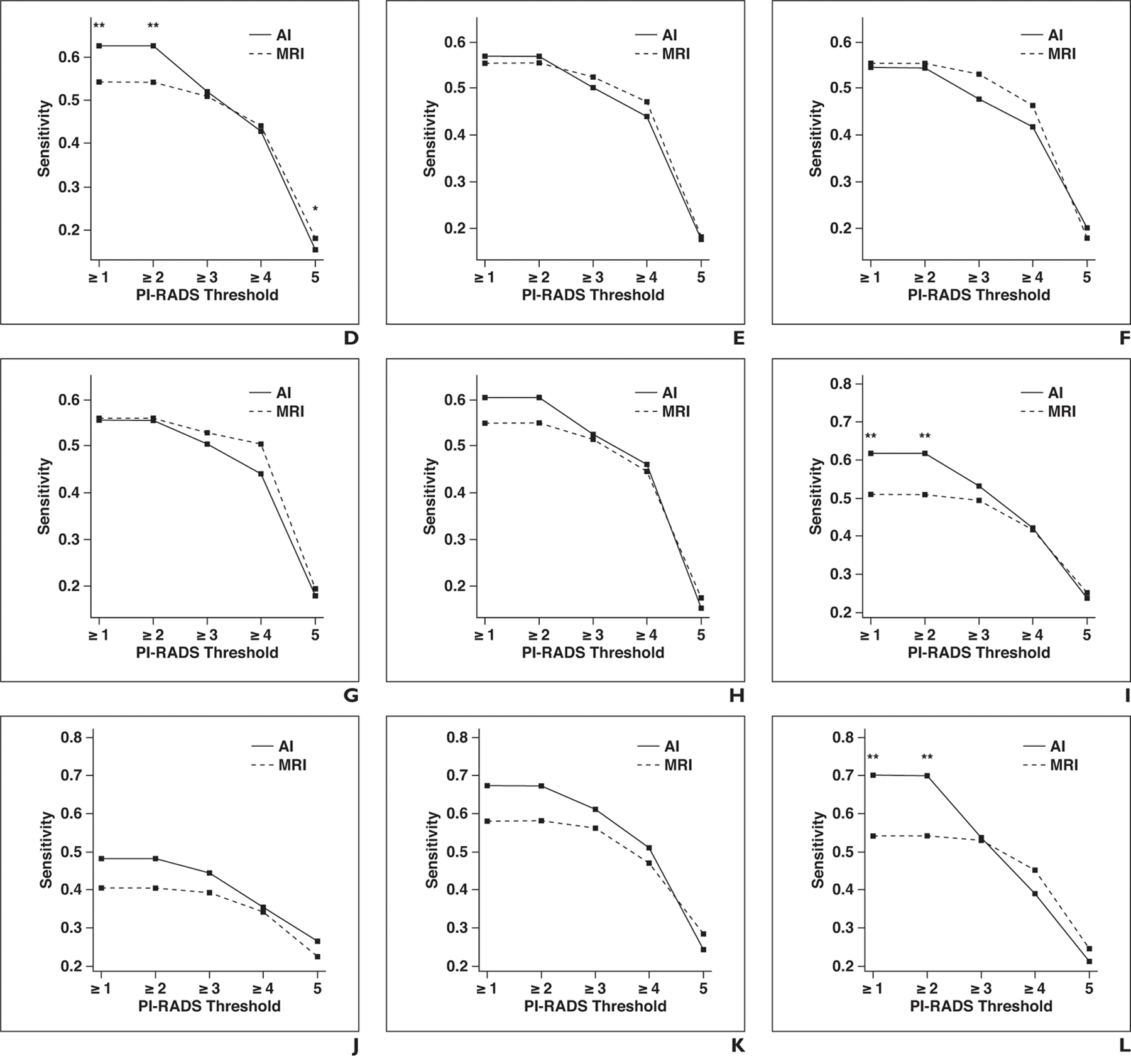
Graphs show sensitivity of artificial intelligence (AI) and MRI for different Prostate Imaging Reporting and Data System (PI-RADS) category thresholds at lesion level for whole prostate, peripheral zone, and transition zone. Asterisk denotes p < 0.05; double asterisk, p < 0.01.
A, Whole prostate, all readers.
B, Whole prostate, readers with low level of experience.
C, Whole prostate, readers with moderate level of experience.
D, Whole prostate, readers with high level of experience.
E, Peripheral zone, all readers.
F, Peripheral zone, readers with low level of experience.
G, Peripheral zone, readers with moderate level of experience.
H, Peripheral zone, readers with high level of experience.
I, Transition zone, all readers.
J, Transition zone, readers with low level of experience.
K, Transition zone, readers with moderate level of experience.
L, Transition zone, readers with high level of experience.
Lesion-level sensitivity and positive predictive value for all PI-RADSv2 thresholds for MRI and AI in the peripheral are shown in Table 3 and in the transition zone in Table 4. There was no statistically significant difference in sensitivity between AI and MRI in the peripheral zone for any PI-RADSv2 threshold. In the transition zone, for the detection of all ground truth lesions (threshold, PI-RADSv2 ≥ 1), sensitivity was significantly higher for AI than for MRI (61.8% vs 50.8%, p = 0.001). In the subgroup of highly experienced readers, sensitivity was also significantly higher for AI than for MRI (70% vs 54.1%, p = 0.003). For lesions considered suspicious on MRI studies (threshold, PI-RADSv2 ≥ 3), there was no significant difference in sensitivity between AI and MRI (53% vs 49.4%, p = 0.238). This was also true among all subgroups of reader experience. The mean numbers of region proposals not corresponding to reportable findings according to PI-RADSv2 guidelines were 2.53 (range, 0–4) for control patients and 1.76 (range, 0–4) for case patients.
TABLE 3:
Diagnosis Performance Statistics on Artificial Intelligence (AI) and MRI for PI-RADS Category Thresholds: Peripheral Zone Lesions
| PI-RADS Threshold for Performance Metric Evaluation | AI |
MRI |
||||||
|---|---|---|---|---|---|---|---|---|
| Overall | Reader Experience Level |
Overall | Reader Experience Level |
|||||
| Low | Moderate | High | Low | Moderate | High | |||
| PI-RADS ≥ 1 | ||||||||
| Sensitivity (%) | 56.8 (51.3–63.1) | 54.5 (46.5–63.0) | 56.6 (48.7–63.0) | 60.5 (54.0–68.1) | 55.4 (49.9–61.7) | 55.4 (47.9–63.1) | 55.9 (49.1–63.5) | 55.1 (47.9–63.7) |
| PPV (%) | 52.1 (47.9–56.5) | 54.1 (47.5–61.4) | 47.7 (41.0–54.2) | 54.6 (48.5–61.4) | 58.7 (54.4–63.1) | 59.1 (52.5–66.1) | 51.6 (45.4–58.2) | 65.3 (58.6–72.8) |
| PI-RADS ≥ 2 | ||||||||
| Sensitivity (%) | 56.8 (51.3–63.1) | 54.5 (46.5–63.0) | 55.6 (48.7–63.0) | 60.5 (54.0–68.1) | 55.4 (49.9–61.7) | 55.4 (47.9–63.1) | 55.9 (49.1–63.5) | 55.1 (47.9–63.7) |
| PPV (%) | 52.5 (48.2–56.9) | 54.1 (47.5–61.4) | 48.3 (41.6–54.8) | 55.0 (49.0–61.6) | 59.3 (55.1–63.7) | 61.0 (54.1–68.2) | 51.6 (45.4–58.0) | 65.3 (58.6–72.8) |
| PI-RADS ≥ 3 | ||||||||
| Sensitivity (%) | 50.1 (44.2–57.1) | 47.6 (39.7–56.5) | 50.4 (43.2–58.4) | 52.5 (44.9–61.5) | 52.4 (47.2–58.7) | 53.0 (45.8–60.8) | 53.0 (46.0–61.0) | 51.3 (44.1–60.4) |
| PPV (%) | 60.4 (55.9–65.3) | 60.7 (53.5–68.6) | 54.1 (46.7–61.3) | 66.6 (60.6–72.7) | 63.4 (59.1–67.8) | 66.1 (58.7–73.8) | 52.2 (45.4–59.1) | 71.9 (65.9–78.3) |
| PI-RADS ≥ 4 | ||||||||
| Sensitivity (%) | 44.0 (38.4–50.7) | 41.9 (34.8–50.0) | 44.1 (36.9–52.2) | 46.0 (39.9–54.3) | 47.2 (41.6–53.5) | 46.4 (38.5–54.7) | 50.5 (43.5–58.7) | 44.6 (38.2–53.2) |
| PPV (%) | 67.5 (62.8–72.5) | 64.2 (55.9–72.9) | 59.5 (51.5–67.5) | 78.8 (72.9–84.9) | 67.8 (62.9–72.5) | 68.6 (60.8–76.4) | 55.1 (48.2–62.3) | 79.5 (72.9–86.4) |
| PI-RADS ≥ 5 | ||||||||
| Sensitivity (%) | 17.8 (13.4–22.8) | 20.0 (13.9–26.5) | 18.0 (12.0–25.0) | 15.3 (10.2–21.7) | 18.2 (13.6–23.4) | 17.8 (11.6–24.6) | 19.3 (13.3–26.7) | 17.4 (11.7–24.5) |
| PPV (%) | 88.6 (82.4–94.1) | 90.8 (82.7–97.9) | 83.1 (72.4–93.3) | 91.9 (82.4–100) | 84.7 (79.0–90.9) | 84.3 (74.2–94.1) | 83.5 (72.4–94.9) | 86.4 (79.6–93.3) |
Note—Values in parentheses are 95% CI. PI-RADS = Prostate Imaging Reporting and Data System, PPV = positive predictive value.
TABLE 4:
Diagnosis Performance Statistics on Artificial Intelligence and MRI for PI-RADS Category Thresholds: Transition Zone Lesions
| PI-RADS Threshold for Performance Metric Evaluation | AI |
MRI |
||||||
|---|---|---|---|---|---|---|---|---|
| Overall | Reader Experience Level |
Overall | Reader Experience Level |
|||||
| Low | Moderate | High | Low | Moderate | High | |||
| PI-RADS ≥ 1 | ||||||||
| Sensitivity (%) | 61.8 (53.9–70.1)a | 48.1 (34.9–61.9) | 67.2 (54.2–80.4) | 70.0 (60.8–80.1)a | 50.8 (42.3–61.2)a | 40.3 (6.3–28.6) | 58.1 (45.3–72.3) | 54.1 (42.7–67.8)a |
| PPV (%) | 34.4 (29.6–39.9) | 39.9 (29.8–50.8) | 28.5 (21.6–37.3) | 34.7 (28.4–41.7) | 57.7 (50.3–64.3) | 61.4 (7.6–44.9) | 53.6 (43.9–63.6) | 58.0 (47.2–68.9) |
| PI-RADS ≥ 2 | ||||||||
| Sensitivity (%) | 61.8 (53.9–70.1)a | 48.1 (34.9–61.9) | 67.2 (54.2–80.4) | 70.0 (60.8–80.1)a | 50.8 (42.3–61.2)a | 40.3 (6.3–28.6) | 58.1 (45.3–72.3) | 54.1 (42.7–67.8)a |
| PPV (%) | 34.5 (29.7–40.0) | 39.9 (29.8–50.8) | 28.5 (21.6–37.3) | 35.0 (28.5–42.1) | 58.2 (50.8–64.8) | 62.2 (7.6–45.7) | 54.3 (44.8–64.4) | 58.0 (47.2–68.9) |
| PI-RADS ≥ 3 | ||||||||
| Sensitivity (%) | 53.0 (44.8–62.8) | 44.3 (32.7–57.4) | 61.1 (48.4–75.4) | 53.5 (41.4–66.6) | 49.4 (41.2–59.8) | 39.1 (6.4–27.2) | 56.1 (43.0–70.8) | 52.9 (41.7–66.7) |
| PPV (%) | 46.2 (40.7–52.9) | 41.3 (30.9–52.9) | 42.1 (34.3–50.7) | 55.2 (45.5–65.4) | 64.6 (58.9–70.7) | 67.7 (5.1–57.6) | 57.8 (47.9–67.7) | 68.4 (58.9–78.5) |
| PI-RADS ≥ 4 | ||||||||
| Sensitivity (%) | 41.8 (33.8–51.1) | 35.3 (25.5–46.4) | 51.0 (38.4–65.5) | 38.9 (28.2–50.8) | 42.0 (34.2–51.5) | 34.1 (5.7–23.7) | 47.0 (34.9–61.4) | 45.0 (33.5–58.3) |
| PPV (%) | 64.9 (57.3–73.1) | 51.7 (38.2–66.2) | 66.5 (55.9–77.4) | 76.5 (64.7–88.4) | 74.6 (68.6–81.0) | 73.6 (5.7–62.6) | 63.2 (51.9–74.3) | 87.1 (79.4–94.6) |
| PI-RADS ≥ 5 | ||||||||
| Sensitivity (%) | 23.9 (16.8–32.4) | 26.4 (16.5–37.0) | 24.2 (14.9–35.3) | 21.1 (11.3–33.1) | 25.0 (17.5–34.5) | 22.5 (5.3–13.3) | 28.3 (18.5–40.1) | 24.4 (13.8–36.6) |
| PPV (%) | 86.6 (79.3–94.4) | 86.7 (76.2–100) | 82.2 (63.9–97.0) | 91.1 (81.5–100) | 86.4 (77.8–93.8) | 86.5 (7.1–70.3) | 81.0 (63.9–96.3) | 91.7 (83.3–100) |
Note—Values in parentheses are 95% CI. AI = artificial intelligence, PI-RADS = Prostate Imaging Reporting and Data System, PPV = positive predictive value.
p < 0.01.
Interreader Agreement
There was moderate interreader agreement in the whole group for AI and MRI with no statistically significant difference (58.7% vs 58.5%, p = 0.966). This was also true of readers with low (55.2% vs 48.5%, p = 0.403) and moderate (55.7% vs 55.5%, p = 0.993) experience levels. Among highly experienced readers, interreader agreement was substantial for AI and MRI with no statistically significant difference (0.644 vs 0.645, p = 0.959).
Image Interpretation Times
The overall reading time was significantly longer for AI than for MRI (4.66 vs 4.03 minutes, p < 0.001). This was particularly pronounced among moderately (5.41 vs 4.68 minutes, p = 0.001) and highly (4.22 vs 3.33 minutes, p < 0.001) experienced readers, but there was no statistically significant difference among readers with a low level of experience (4.33 vs 4.1 minutes, p = 0.289).
Discussion
In this study an AI detection system entailing region-based attention mapping showed little or no improvement over conventional interpretation of prostate MRI across multiple readers of various experience levels. The notable exception was overall improvement in the detection of transition zone lesions, which are more difficult to diagnose. This result agrees with those of other AI studies, which to date have not shown dramatic improvements in performance over conventional interpretation. Interestingly, the AI system did not improve interreader variability or improve the performance of readers with a low level of experience, which are features commonly touted for AI systems.
This study revealed some interesting findings in subset analysis. For instance, among highly experienced readers, sensitivity was significantly higher for AI than for MRI for all lesions; this difference was not observed for lesions deemed suspicious for prostate cancer with MRI. In other words, the AI system increased the sensitivity of highly experienced radiologists in detecting invisible (PI-RADS 1) and low-category (PI-RADS 2) lesions, whereas it was not as contributory for clearly visible lesions (PI-RADS ≥ 3).
One possible explanation for the more pronounced effect on highly experienced readers could be the different output format of our AI system. Although our previous and most other AI systems use color-coded cancer probability maps, we chose an attention-based mapping box to decrease distraction and subjectivity caused by background noise and false-positive lesions on AI maps and focus the reader’s attention on the areas of highest likelihood of cancer. It is possible that highly experienced readers benefitted most from this approach because they were more confident in detecting cancer-suspicious lesions and could spend additional time on the regions highlighted by the AI derived boxes, whereas less experienced readers might be less confident and not weight the AI data as strongly.
The AI system performed differently in different zones of the prostate. The peripheral and transition zones have very different radiologic and histopathologic properties. As a consequence, the PI-RADS consensus guidelines recommend different criteria for the assignment of risk categories in the peripheral and transition zones [12]. The transition zone usually has a heterogeneous signal-intensity pattern on T2-weighted images, especially in patients with benign prostatic hyperplasia. This complicates the detection of prostate cancer lesions, which therefore can be easily overlooked. As a result, the sensitivity of mpMRI in general is lower for transition zone than for peripheral zone lesions [17]. In the subgroup analysis of peripheral and transition zone lesions in our study, the AI system had significantly higher sensitivity for all MRI-detected transition zone lesions than did mpMRI. This may represent an important contribution of this detection system.
Current AI systems are not fully automated detection systems but rather adjunct tools to aid radiologists reading prostate mpMRI studies. Therefore, the AI information along with prostate mpMRI findings can be considered an additional parameter potentially increasing complexity for no benefit. Our study did not show significant improvement in interreader agreement among readers. Reading time was slightly longer (mean, 40 seconds) with the AI detection system. This is understandable because the nature of an attention box is that it requires additional time to evaluate. Some AI systems reduce the time needed to diagnose. In a study by Greer et al. [18], both interreader variability and readout times improved when AI was used. In that study, however, the mpMR images were acquired at one site with one set of acquisitions, whereas the current study included MRI studies from multiple institutions. It is possible that such a heterogeneous collection of MRI data may require more time to evaluate even with an AI detection system. Additionally, spending extra time to carefully search for suspicious lesions within the four attention boxes in each patient may also have increased readout time.
Limitations
Our study had several limitations. First, the output of the AI detection system always presented four attention boxes to the reader even if there were no lesions. This was done because PI-RADS allows evaluation of up to four lesions and there was no method for varying the number of boxes for each case. This likely resulted in more false-positive readings, resulting in significantly lower specificity on the patient level compared with prior AI detection systems.
Second, the training of the algorithm was based on a fairly small patient population. This might have negatively affected the performance of the model because larger and more diverse patient populations improve generalizability of classification algorithms.
Third, our classification algorithm was based on a random forest classifier. With advances in computational resources, big data, and more sophisticated deep neural network algorithms, deep learning is gaining popularity in medicine. In particular, convolutional neural networks appear to be superior to classic machine learning techniques and other deep neural network architectures in performance and generalization in imaging-related tasks [19–21]. As a result, we are currently working on procuring and annotating larger imaging datasets and developing deep neural network architectures for designing a stronger prostate cancer AI detection system.
Conclusion
The AI detection system had significantly higher sensitivity than mpMRI for the detection of transition zone lesions, especially those not visible to readers using the raw images alone. Overall, there was no significant gain from the AI detection system compared with MRI alone, and it did not improve the performance of readers with a low experience level or reduce interreader variability. AI did, however, improve the performance of radiologists in evaluating transition zone lesions. These results suggest a need for further work on deep learning convolutional neural networks in larger datasets to improve the performance of radiologists interpreting mpMRI of the prostate.
Acknowledgments
Supported in whole or in part by federal funds from the National Cancer Institute, National Institutes of Health (contract HHSN261200800001E).
Footnotes
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018; 68:7–30 [DOI] [PubMed] [Google Scholar]
- 2.Hricak H, Williams RD, Spring DB, et al. Anatomy and pathology of the male pelvis by magnetic resonance imaging. AJR 1983; 141:1101–1110 [DOI] [PubMed] [Google Scholar]
- 3.Hamoen EHJ, de Rooij M, Witjes JA, Barentsz JO, Rovers MM. Use of the Prostate Imaging Reporting and Data System (PI-RADS) for prostate cancer detection with multiparametric magnetic resonance imaging: a diagnostic meta-analysis. Eur Urol 2015; 67:1112–1121 [DOI] [PubMed] [Google Scholar]
- 4.Tan N, Margolis DJ, Lu DY, et al. Characteristics of detected and missed prostate cancer foci on 3-T multiparametric MRI using an endorectal coil correlated with whole-mount thin-section histopathology. AJR 2015; 205:[web]W87–W92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le JD, Tan N, Shkolyar E, et al. Multifocality and prostate cancer detection by multiparametric magnetic resonance imaging: correlation with whole-mount histopathology. Eur Urol 2015; 67:569–576 [DOI] [PubMed] [Google Scholar]
- 6.Serrao EM, Barrett T, Wadhwa K, et al. Investigating the ability of multiparametric MRI to exclude significant prostate cancer prior to transperineal biopsy. Can Urol Assoc J 2015; 9:E853–E858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lay N, Tsehay Y, Greer MD, et al. Detection of prostate cancer in multiparametric MRI using random forest with instance weighting. J Med Imaging (Bellingham) 2017; 4:024506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaur S, Lay N, Harmon SA, et al. Can computer-aided diagnosis assist in the identification of prostate cancer on prostate MRI? A multi-center, multi-reader investigation. Oncotarget 2018; 9:33804–33817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cochran WG, Cox GM. Experimental designs, 2nd ed. New York, NY: Wiley, 1992 [Google Scholar]
- 10.Greer MD, Lay N, Shih JH, et al. Computer-aided diagnosis prior to conventional interpretation of prostate mpMRI: an international multi-reader study. Eur Radiol 2018; 28:4407–4417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Criminisi A, Shotton J, Robertson D, Konukoglu E. Regression forests for efficient anatomy detection and localization in CT studies. In: Medical computer vision: recognition techniques and applications in medical imaging. Berlin, Germany: Springer, 2011:106–117 [Google Scholar]
- 12.American College of Radiology. PI-RADStm: Prostate Imaging and Reporting and Data System, version 2. Reston, VA: American College of Radiology, 2015 [Google Scholar]
- 13.Berbaum KS, Krupinski EA, Schartz KM, et al. Satisfaction of search in chest radiography. Acad Radiol 2015; 22:1457–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Epstein JI, Egevad L, Amin MB, Delahunt B, Srigley JR, Humphrey PA.; Grading Committee. The 2014 International Society of Urological Pathology (ISUP) consensus conference on Gleason grading of prostatic carcinoma: definition of grading patterns and proposal for a new grading system. Am J Surg Pathol 2016; 40:244–252 [DOI] [PubMed] [Google Scholar]
- 15.Bandos AI, Obuchowski NA. Evaluation of diagnostic accuracy in free-response detection-localization tasks using ROC tools. Stat Methods Med Res 2019; 28:1808–1825 [DOI] [PubMed] [Google Scholar]
- 16.Shih JH, Greer MD, Turkbey B. The problems with the kappa statistic as a metric of interobserver agreement on lesion detection using a third-reader approach when locations are not prespecified. Acad Radiol 2018; 25:1325–1332 [DOI] [PubMed] [Google Scholar]
- 17.Li W, Xin C, Zhang L, Dong A, Xu H, Wu Y. Comparison of diagnostic performance between two prostate imaging reporting and data system versions: a systematic review. Eur J Radiol 2019; 114:111–119 [DOI] [PubMed] [Google Scholar]
- 18.Greer MD, Brown AM, Shih JH, et al. Accuracy and agreement of PIRADSv2 for prostate cancer mpMRI: a multireader study. J Magn Reson Imaging 2017; 45:579–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ting DSW, Cheung CYL, Lim G, et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA 2017; 318:2211–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ehteshami Bejnordi B, Veta M, Johannes van Diest P, et al. ; The CAMELYON16 Consortium. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 2017; 318:2199–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winkler JK, Fink C, Toberer F, et al. Association between surgical skin markings in dermoscopic images and diagnostic performance of a deep learning convolutional neural network for melanoma recognition. JAMA Dermatol 2019. Aug 14 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]


