FIGURE 2.
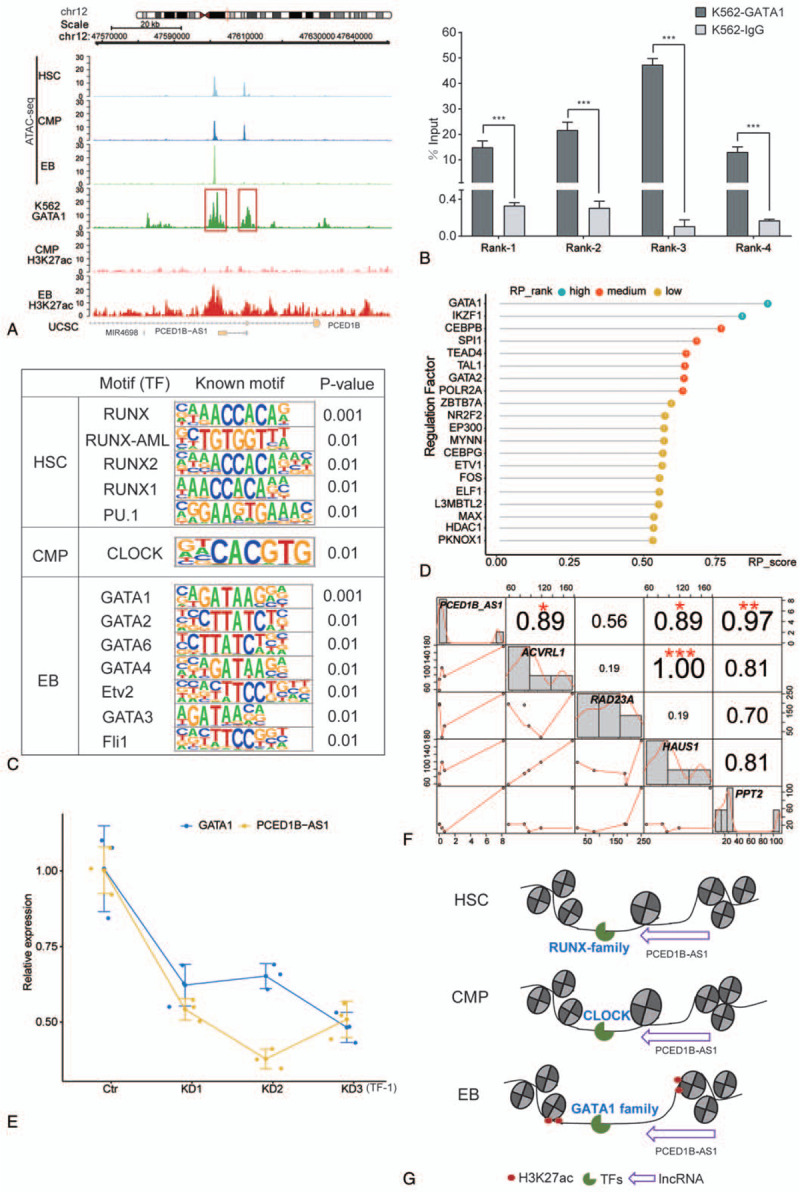
The association between lncRNA PCED1B-AS1 and chromatin accessibility during erythroid differentiation. (A) Dynamic profile of chromatin accessibility and histone modification of H3K27ac around PCED1B-AS1 loci during erythroid differentiation. (B) Confirmation of physical binding of GATA1 around PCED1B-AS1 loci by ChIP-qPCR in K562 cells. Rank1 and Rank2 are from the red box on the left side in (A) picture, and Rank3 and Rank4 are from the red box on the right side in (A). The bar graphs represent the average of percentage input (mean ± SD) from three independent ChIP experiments. (C) Known motif identification of peaks within 20 kb of PCED1B-AS1. (D) Regulation factors of PCED1B-AS1 within its 100 kb genomic regions in EB. (E) Relative expression of PCED1B-AS1 in TF-1 cells with GATA1 knockdown by siRNA. (F) Correlation between PCED1B-AS1 with its potential target genes. (G) Model of PCED1B-AS1 participates in erythroid differentiation via dynamic chromatin remodeling involving GATA1.
