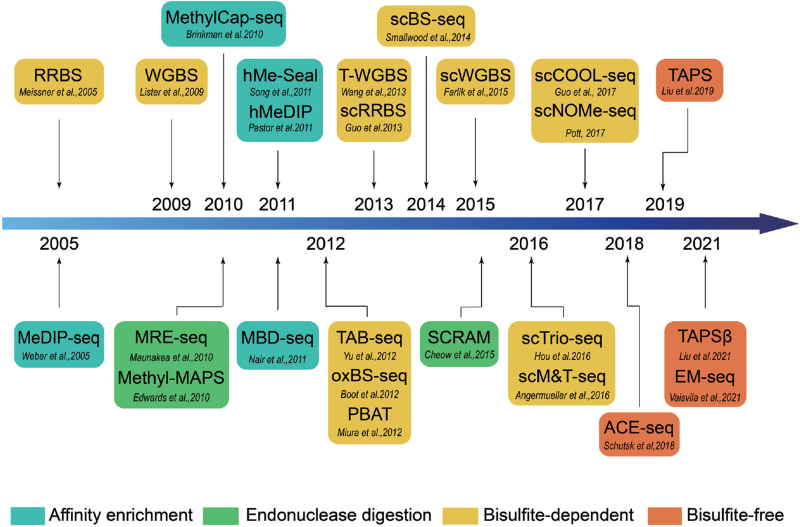
Figure 1.
Evolution of NGS-based techniques applied to DNA methylation profiling. Different background colors indicate different categories of DNA methylation measuring methods (bottom). ACE-seq: APOBEC-coupled epigenetic sequencing, EM-seq = Enzymatic Methyl-seq, hMeDIP = hydroxymethylated DNA immunoprecipitation, hMe-Seal = 5hmC-selective chemical labeling method, MBD-seq = methyl-CpG-binding domain protein sequencing, MeDIP-seq = methylated DNA immunoprecipitation sequencing, Methyl-MAPS = methylation mapping analysis by paired-end sequencing, MRE-seq = methylation sensitive restriction enzyme sequencing, oxBS-seq: oxidative bisulfite sequencing, PBAT = post-bisulfite adaptor tagging, RRBS = reduced representation bisulfite sequencing, scBS-seq = single cell bisulfite sequencing, scCOOL-seq = single-cell Chromatin Overall Omic-scale Landscape Sequencing, scM&T-seq = single-cell methylome and transcriptome sequencing, scNOMe-seq = single-cell Nucleosome Occupancy and Methylome Sequencing, SCRAM = single-cell restriction analysis of methylation, scRRBS = single-cell reduced representation bisulfite sequencing, scTrio-seq = single-cell triple omics sequencing, scWGBS = single cell whole genome bisulfite sequencing, TAB-seq = Tet-assisted bisulfite sequencing, TAPS = TET-assisted pyridine borane sequencing, TAPSβ = TAPS with β-glucosyltransferase (β-GT) blocking, T-WGBS = Tagmentation based WGBS, WGBS = whole-genome bisulfite sequencing.