Figure 1.
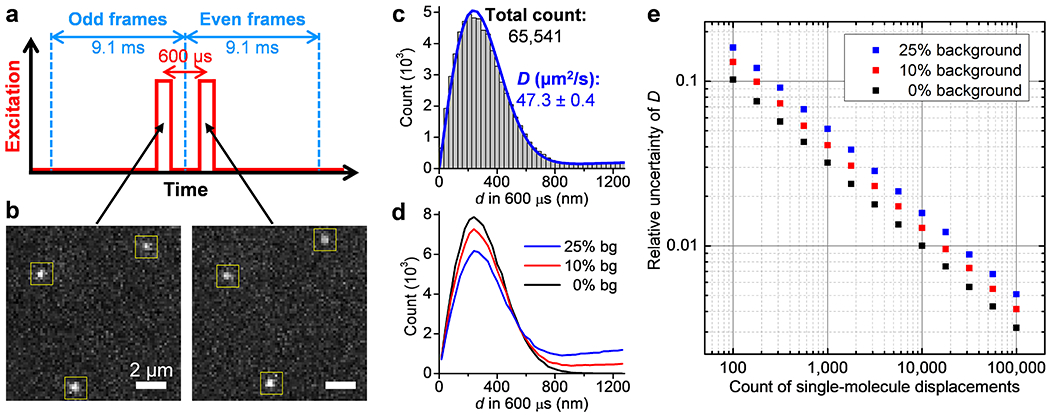
High-throughput statistics of unhindered single-molecule displacements. (a) Schematics: tandem excitation pulses of 300 µs duration are applied at Δt = 600 µs center-to-center separations across paired frames of the recording EM-CCD camera. (b) Example single-molecule images captured in the paired frames, for Cy3B-labeled catalase freely diffusing in the PBS buffer, which enabled the determination of the transient displacements d of the 3 molecules marked by the yellow boxes in the Δt = 600 µs time window. (c) Histogram: distribution of the 65,541 transient single-molecule displacements collected in 9 min by repeating the tandem excitation scheme 30,000 times. Blue line: MLE yielding a diffusion coefficient D of 47.3±0.4 µm2/s (95% confidence interval). (d) Example distributions of 100,000 simulated displacements, of which 0%, 10%, and 25% are backgrounds from molecules randomly entered the vicinity while the rest are 600-µs displacements with D = 50 µm2/s. (e) Relative uncertainty of D (σD/D) from MLE results of the simulated data at different levels of backgrounds, as a function of the counts of displacements. The standard deviations σD were calculated from the differences between 5,000 rounds of simulations under each condition.
