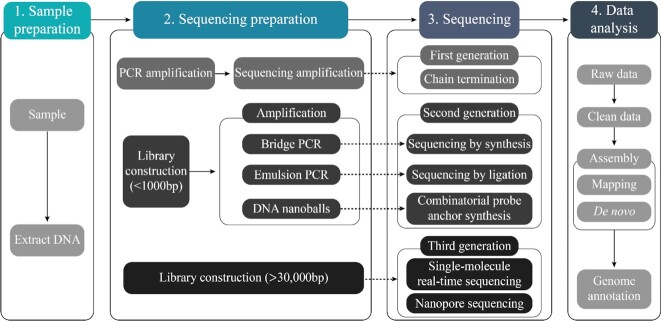
Figure 2:
Schematic representation of the next-generation sequencing pipeline for genomic assembly. (1) Multiple companies have marketed sequencing platforms for genomic and transcriptomic studies, the most commonly used being Illumina (left), PacBio (middle), and Nanopore (right). (2) The 3 platforms differ in read length and accuracy of their generated sequences. Whilst Illumina sequencing generally yields short reads with low error rates, Nanopore sequences are substantially longer (≤2 Mb), yet subject to frequent sequencing errors. Last, PacBio generates sequences with lengths and error rates in between the 2 other platforms. (3) After sequencing, reads are computationally processed and assembled into contigs, which in turn (4) serve as the building blocks for scaffolds.The scaffolds are then aligned and annotated to produce the complete target genome.