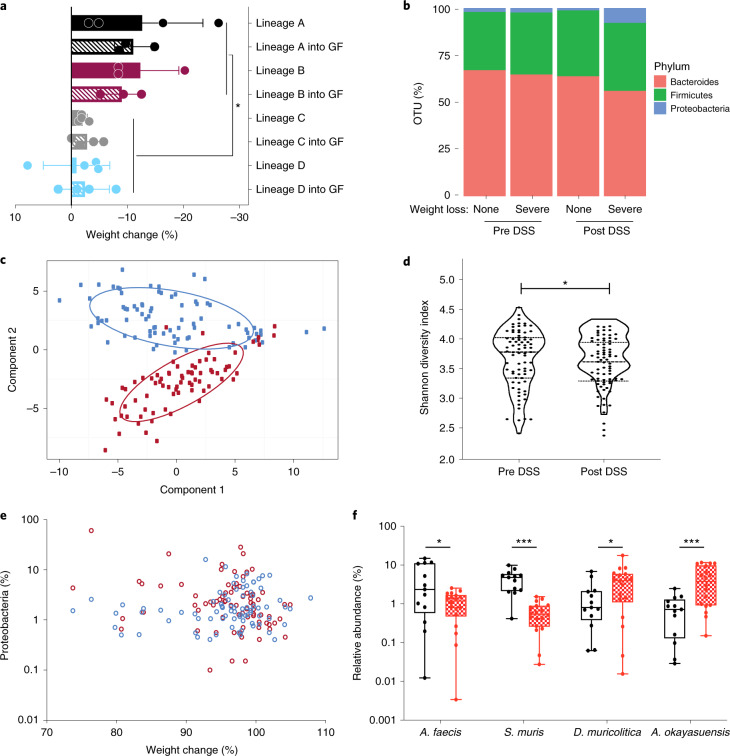
Fig. 2. Discovery of candidate bacterial taxa associated with DSS disease variability.
a, Comparison of weight change observed in conventional SPF mice (lineages A, B, C and D, solid colors) and germ-free (GF) mice colonized by oral gavage (lineages A, B, C and D into GF, hatched colors) after 10-day DSS challenge (P = 0.0002, two-tailed t-test with Welch’s correction; data plotted as mean ± s.d.; n = 4 (lineages A, C, C + GF, D and D + GF) and n = 3 (lineages B, B + GF, A + GF)). b, Distribution of Bacteroides (red), Firmicutes (green) and Proteobacteria (blue) in pre- and post-DSS samples exhibiting asymptomatic or >5% weight loss and inflammation. c, Principal component analysis of 16 S rRNA amplicon sequencing profiling identifies a clear difference between pre-DSS samples (blue) and post-DSS samples (red), regardless of lineage. d, Alpha diversity using Shannon diversity index grouped by pre- and post-DSS samples (*P = 0.0357, paired t-test; n = 77). e, Relationship between weight change and relative abundance of Proteobacteria pre DSS challenge (blue; Spearman correlation, 0.196) and post DSS (red; Spearman correlation, −0.289). f, Relative abundance of A. faecis, A. okayasuensis, D. muricolitica and S. muris before challenge with DSS amongst samples from mice that experienced >5% weight loss (red) or no weight loss (black) (P = 0.0264, 0.0006, 0.0351, 0.0010, two-tailed t-test; n = 18 and n = 13 independent animals, respectively. Minima (25th percentile) median (75th percentile) and maxima are shown. *P < 0.05; ***P ≤ 0.001).