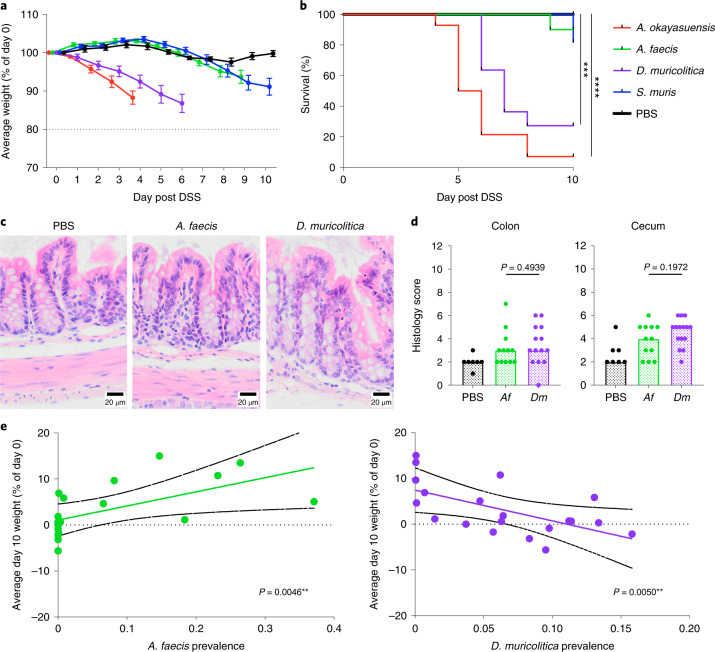
Fig. 3. Phenotypic analysis of mice monocolonized with candidate bacterial taxa.
a,b, Average weight loss/gain (a) and survival (b) in gnotobiotic mice precolonized with either A. okayasuensis (red; n = 14 animals, P = 0.00001), A. faecis (green; n = 10 animals, P = 0.31731), D. muricolitica (purple; n = 10 animals, P = 0.00074) or S. muris (blue; n = 11 animals, P = 0.16653) and in germ-free controls (PBS; black; n = 10 animals) in response to oral DSS challenge. All survival curves were compared to PBS control by log-rank (Mantel–Cox) test, with median and interquartile range plotted. c,d, Representative colon images (400×) from two experiments, each with between three and seven mice per group (c), and histological scores from H&E staining of midcolon (d) (P = 0.4939, Mann–Whitney test) and cecum (P = 0.1972, Mann–Whitney test) from either control germ-free mice (PBS) or mice monocolonized with either A. faecis (Af, n = 12) or D. muricolitica (Df, n = 14) and challenged with oral DSS for 7 days. Average weight loss curves were censored after the first mouse in each group reached 80% of baseline weight. e, Day 10 weight loss relative to day 0 starting weight in 20 mice, showing a positive correlation (green line) between weight change and prevalence in A. faecis (R2 = 0.3679, P = 0.0046) and negative correlation (purple line) between weight change and prevalence in D. muricolitica (R2 = 0.3626, P = 0.005). Solid black lines indicate 95% confidence interval. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.