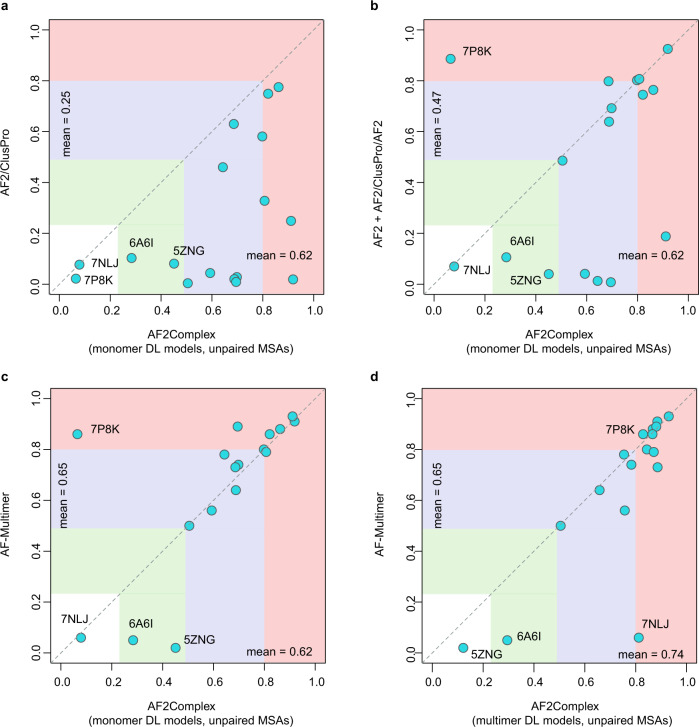
Fig. 3. Comparison of AF2Complex and three alternative approaches on the CP17 set.
The coordinates of the circles correspond to the DockQ scores42 of the top overall models from each approach versus AF2Complex. a AF2 models docked by ClusPro22. b Docking models refined by AF2, plus additional complex models obtained by running AF2 on paired MSAs according to Ref. 22. (c, d) AlphaFold-Multimer35. The AF2 deep learning models trained for the prediction of monomeric protein structures, denoted as “monomer DL models”, were employed by AF2Complex in (A–C), and the AF-Multimer deep learning models, denoted as “multimer DL models”, were applied with AF2Complex in (d). All MSA inputs to AF2Complex are unpaired as described in Methods. Vertical and horizontal blocks represent the regions of incorrect (white), acceptable (green), medium (blue), and high-quality (red) complex models according to the DockQ score. The four most challenging targets are marked by their four-letter PDB accession codes.