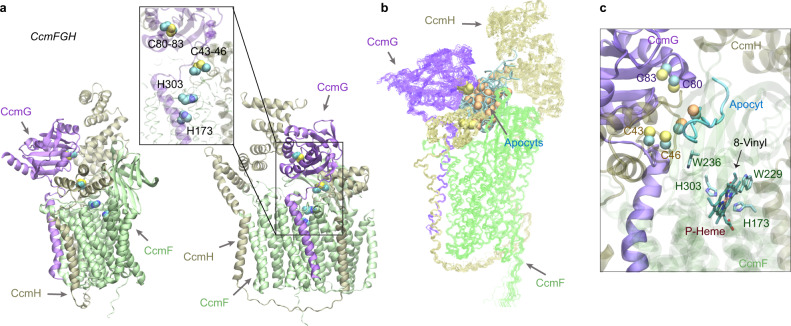
Fig. 8. Structural models of the CcmFGH complex with and without apo-cytochrome c substrates (apocyts) from E. coli.
a Two views of a top model of CcmFGH are shown in the same orientations as the two views in Fig. 7a, respectively. The CcmH N-terminal domain moves closer to the heme-binding sites of CcmF, leaving space to accommodate CcmG that now binds CcmF with CcmH. Critical cysteines of the CXXC motifs of CcmG (Cys80 and Cys83) and H (Cys43 and Cys46), and the P-heme-binding histidines (His173 and His303) of CcmE are shown in the vdW representations in the inset. b Superposition of 22 AF2Complex models of the CcmEFH and apocyt acceptors. CcmFGH complexes are shown in lines, and apocyts are shown as cyan tubes. All apocyts are found within the same groove formed by the three Ccm proteins. The superposition used the backbone atoms of CcmFGH as the reference. The sulfur atoms from the CXXC motifs of apocyts are shown in orange spheres to differentiate from those of CcmGH. c A heme molecule computationally docked to the P-heme binding site in CcmF using one of the models shown in (b). One of the Cys residues of apocyt is found within 4 Å from Cys46 of CcmH. The distance between the other Cys residue of apocyt and the 8-vinyl group of the heme is about 16 Å.