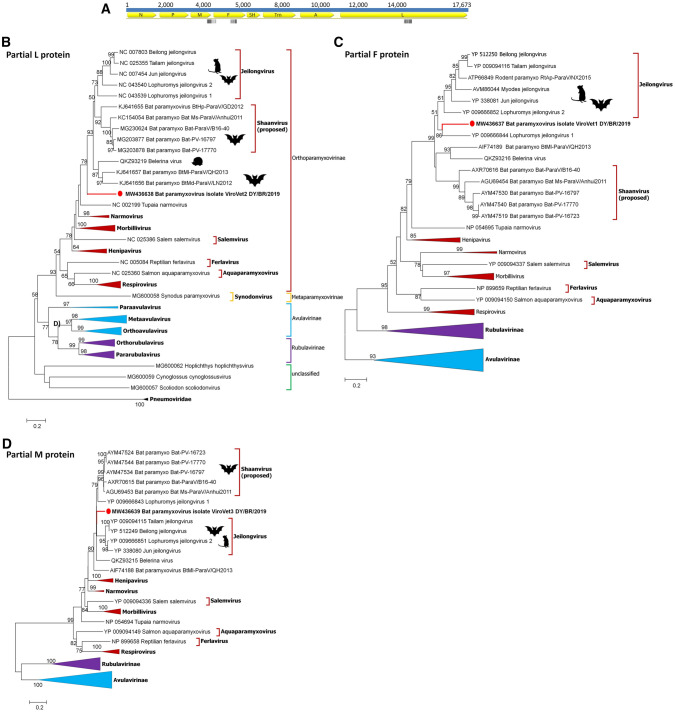
Fig. 5.
a Mapping of bat paramyxovirus contigs ViroVet1, 2, and 3 in the complete genome of a reference bat paramyxovirus (MZ328288.1); b partial L; c partial F; and d partial M amino acid phylogenetic trees of Paramyxoviridae reference sequences and unclassified “bat paramyxovirus”. For the partial L tree, sequences of the Pneumoviridae family were included as outgroups. There were 86, 75, and 74 amino acid sequences, respectively, and 148 positions in the final dataset. The sequences were analyzed through the Maximum likelihood method with LG + G + I model. Analyses were conducted with 1000 bootstrap replicates. Bootstrap values higher than 50% are shown. The sequences detected in the present study were highlighted with circles