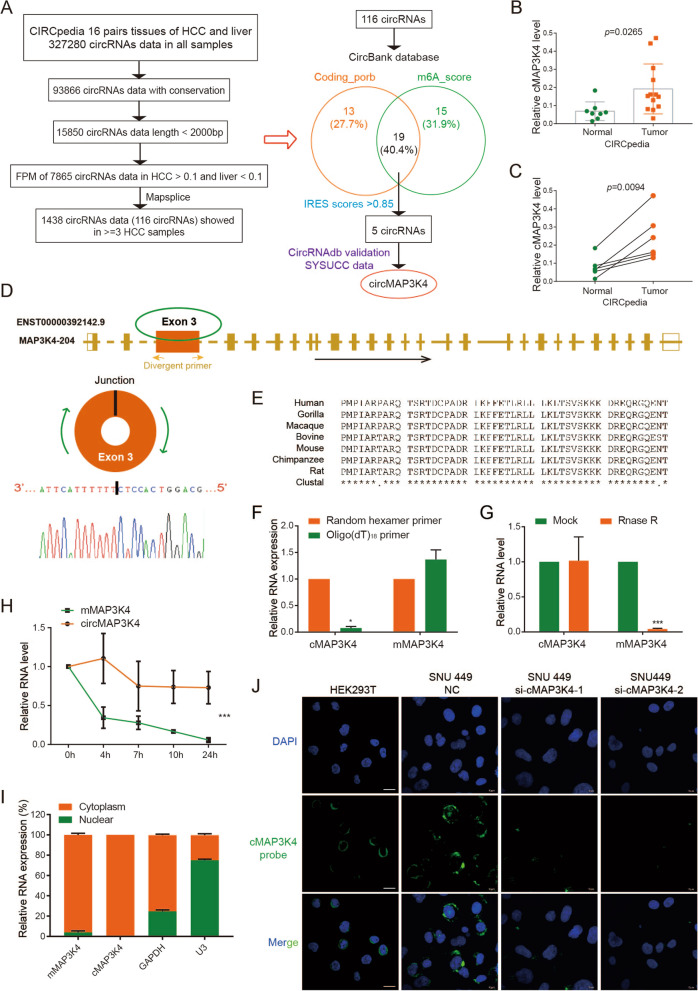
Fig. 1.
Selection and validation of circMAP3K4. A Flowcharts showing the selection of circRNAs overexpressed in HCC with coding potential. B CircMAP3K4 expression in HCC and liver tissues from the CIRCpedia database. Tumor samples, n = 13; liver samples, n = 8. C CircMAP3K4 expression in paired HCC and adjacent liver tissues from the CIRCpedia database (n = 6). D The location of circMAP3K4 in the human genome (upper panel), and circMAP3K4 sketch map. CircMAP3K4 expression detected by RT-qPCR followed by Sanger sequencing using divergent primers. E CircMAP3K4 interspecies sequence conservation. Homo sapiens, Human; Gorilla gorilla, Gorilla; Macaca mulatta, Macaque; Bovini, Bovine; Mus musculus, Mouse; Pan troglodytes, Chimpanzee; Rattus norvegicus, Rat. F–H RT–qPCR analysis of circMAP3K4 expression after amplification with oligo (dT)18 primers, RNase R treatment and actinomycin D treatment, respectively. I CircMAP3K4 distribution detected by cytoplasmic and nuclear RNA fractionation. GAPDH and U3 were employed as positive controls in the cytoplasm and nucleus, respectively. J Fluorescence in situ hybridization showed the predominant cytoplasmic distribution of circMAP3K4 (Scale bar, 20 μm). All experiments were repeated at least three times. Data are shown as mean ± standard deviation (SD), * p < 0.05, ** p < 0.01, and *** p < 0.001 in two-way ANOVA (E) and t-test (C, D). cMAP3K4, circMAP3K4; mMAP3K4, MAP3K4 mRNA