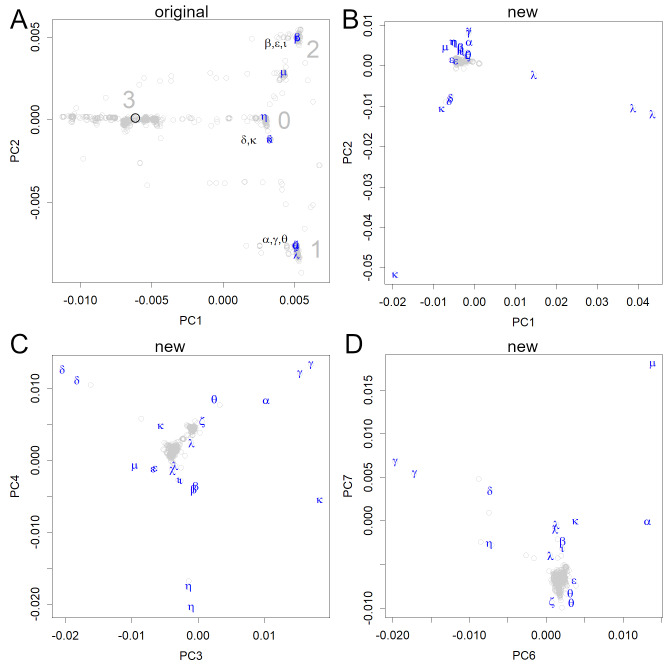
Figure 3. PCA locations of variants of interest to the WHO.
(A) The variants are shown in the axes obtained from the original strain; the sequence data were multiplied with the unitary matrix V obtained by SVD. The variants appeared in groups 0, 1, and 2; many of them belonged to three small groups (the variants, Pango lineages, and earliest documented samples). α: alpha, B.1.1.7, England; β: beta, B.1.351, South Africa; γ: gamma, P.1, Brazil; δ: delta, B.1.617.2, India; ɛ: epsilon, B.1.427&429, USA; ζ: zeta, P.2, Brazil; η: eta, B.1.525, multiple countries; θ: theta, P.3, the Philippines, ι: iota, B.1.526, USA; κ: kappa, B.1.617.1, India; λ: lambda, C.37, Peru, μ: mu, B.1.621, Colombia. (B–D) All data were shown on the axes obtained from the new variants. Each axis shows the features of some variants but does not reveal the initial infection routes.