Figure 1:
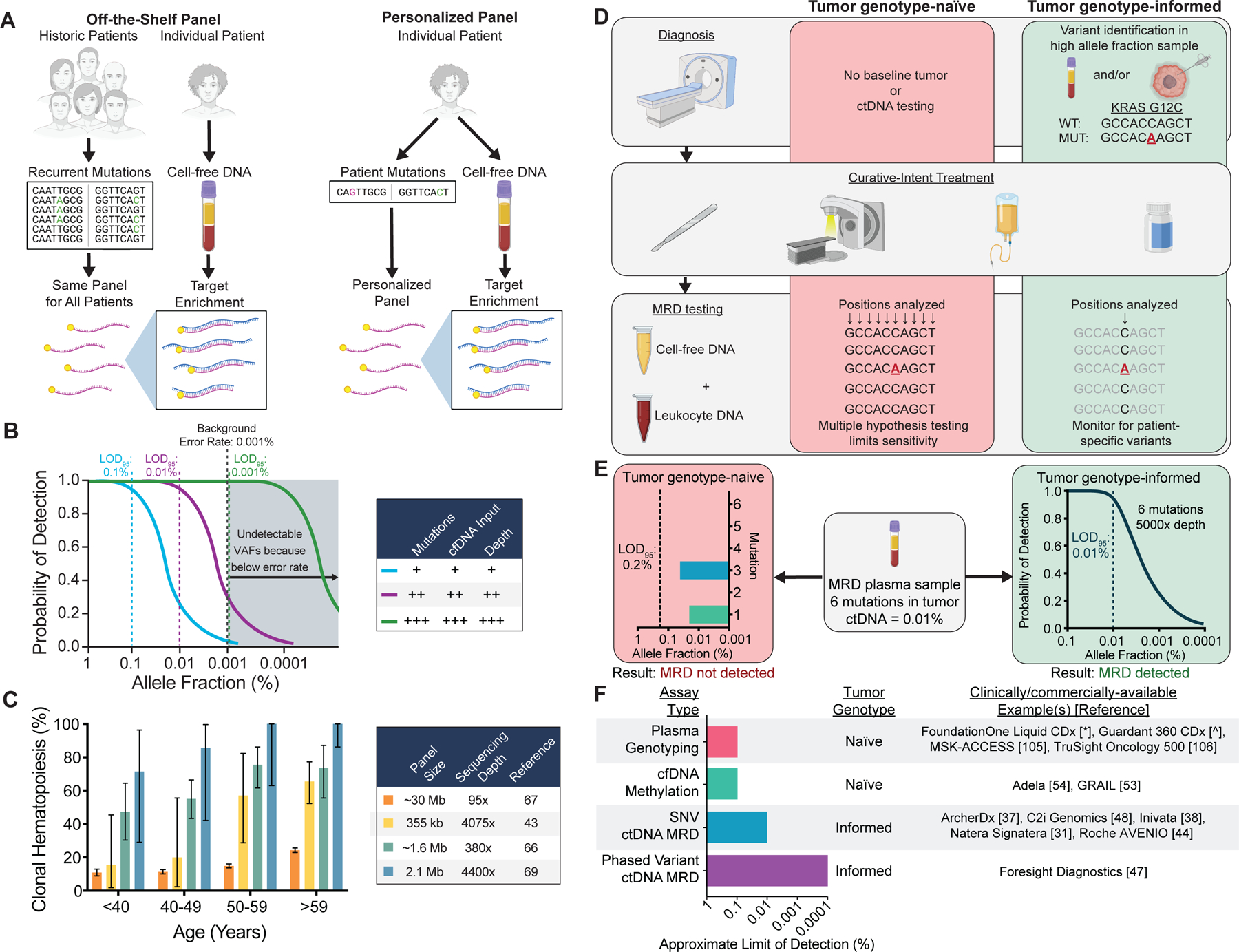
Technical approaches for ctDNA MRD detection and factors affecting assay sensitivity and specificity. A) Schematic comparing assays using off-the-shelf versus personalized sequencing panels. Off-the-shelf panels are designed to cover recurrently mutated genes in the cancer type(s) of interest. The same panel is applied to tumor tissue and plasma of every patient, and personalization is achieved by bioinformatically considering only the positions mutated in the matched tumor . Personalized panels are designed to cover patient-specific mutations identified through sequencing of their tumor DNA. In this approach, personalization is achieved by every patient having a unique panel. B) Factors affecting the probability of ctDNA detection. Increasing the number of mutations tracked, the sequencing depth at mutant positions, or the cfDNA input can increase the probability of detection. Technical background from sources such as polymerase errors and oxidative damage sets the lower limit of detection due to an inability to distinguish artifacts from true tumor variants. Limit of detection with 95% probability = LOD95. C) Prevalence of at least 1 clonal hematopoiesis variant detected in plasma as a function of the sequencing panel size and sequencing depth in published studies (46,75–77). Error bars represent binomial 95% confidence intervals. D) Schematic of tumor genotype-informed versus tumor genotype-naïve ctDNA analysis. In genotype-naïve analysis, the MRD sample is interrogated at all sequenced genomic positions, leading to reduced sensitivity due to multiple hypothesis testing. In tumor genotype-informed analysis, variants are identified from tumor tissue or pre-treatment plasma samples with high tumor allele fraction. Only patient-specific variants are monitored in the MRD sample. In both cases, genotyping DNA from leukocytes in the peripheral blood can improve specificity by identifying variants stemming from clonal hematopoiesis. E) Comparison of the LODs of tumor genotype-naïve and tumor genotype-informed ctDNA analysis at a median sequencing depth of 5000× for a patient with 6 tumor mutations and a ctDNA allele fraction of 0.01%. Due to multiple hypothesis testing with tumor genotype-naïve analysis, the LOD95 is 0.2%, and no mutations are detected above background despite mutations with allele fractions of 0.02% (1/5000 molecules) and 0.04% (2/5000 molecules) being present in the sample. In contrast, in the same sample tumor genotype-informed analysis at 5000× depth is associated with an LOD95 of 0.01% (approximated by a binomial distribution), and therefore ctDNA MRD has a 95% chance of being detected. F) Summary of assay types, tumor genotyping requirements, and approximate LODs (i.e. analytical sensitivity) for commercially/clinically available ctDNA analysis tests. Since no data comparing these methods on the same samples are available, the approximate limit of detection at which 95% of samples would be expected to be called positive for each group of assays is estimated from published manuscripts and/or conference abstracts and rounded to the nearest log (31,37,38,44,47,48,53,54,105,106).
* = http://www.accessdata.fda.gov/cdrh_docs/pdf19/P190032B.pdf
^ = https://www.accessdata.fda.gov/cdrh_docs/pdf20/P200010B.pdf
