Figure 3: Vegfc signaling stimulates emilin2a expression to promote coronary endothelial cell proliferation in regenerating zebrafish hearts.
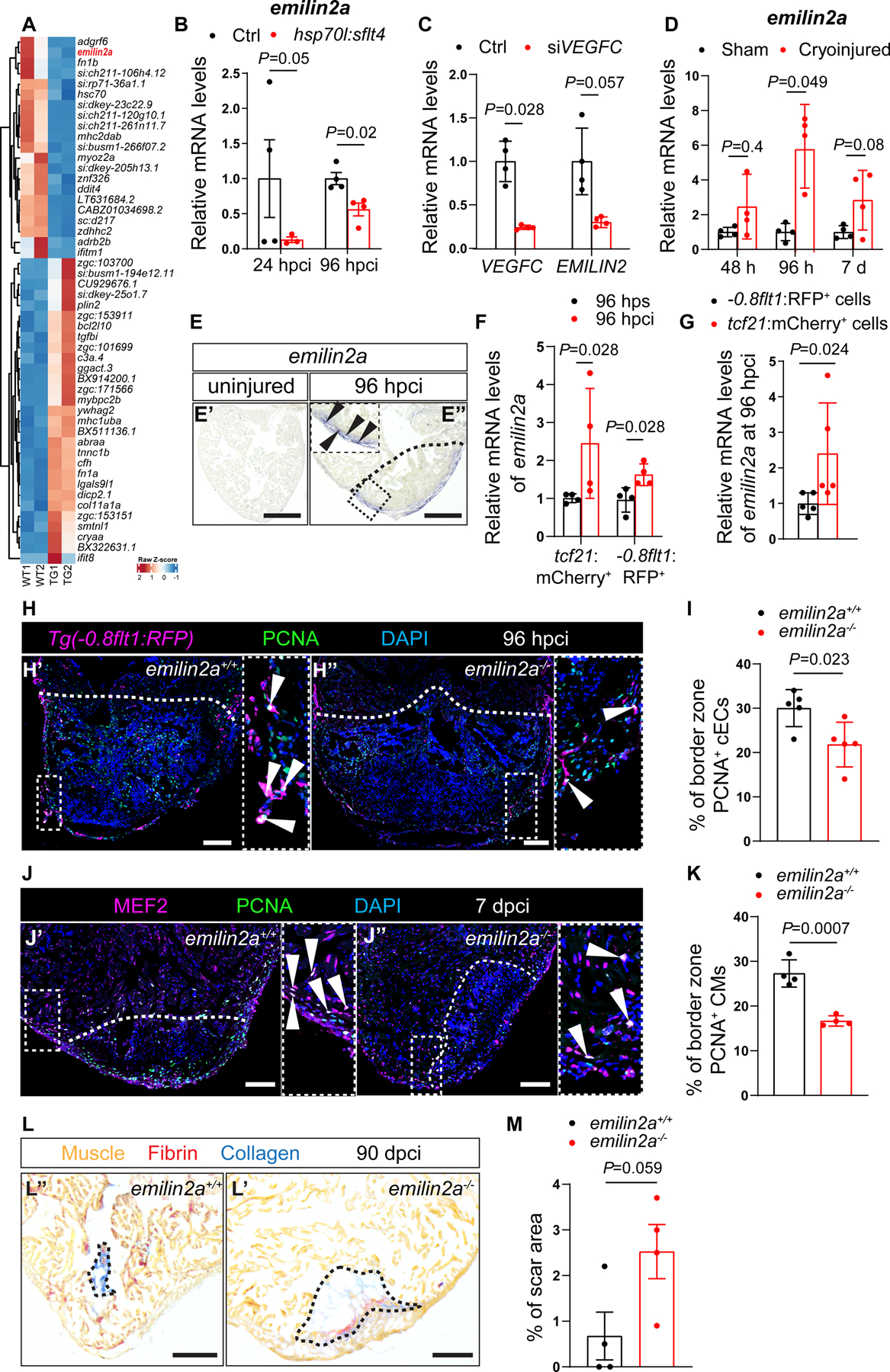
A. Heat map showing differentially expressed genes in non-transgenic sibling (WT) versus Tg(hsp70l:sflt4) hearts at 24 hpci. Genes are broadly divided into two clusters: one that contains genes that are upregulated in Tg(hsp70l:sflt4) ventricles and the other that contains genes that are downregulated in Tg(hsp70l:sflt4) ventricles. B. RT-qPCR analysis of emilin2a mRNA levels at 24 and 96 hpci in Tg(hsp70l:sflt4) ventricles normalized to non-transgenic sibling (Ctrl) hearts (n=4). C. RT-qPCR analysis of VEGFC and EMILIN2 mRNA levels in HUVECs after siVEGFC treatment (n=4) in comparison with scrambled control (n=4). D. RT-qPCR analysis of emilin2a mRNA levels at 48 and 96 hpci and 7 dpci in injured tissue normalized to sham-operated hearts (n=4); h: hpci, d: dpci. E. in situ hybridization for emilin2a expression on sections of uninjured (E’) and 96 hpci (E”) hearts. Arrowheads point to emilin2a expression. F. RT-qPCR analysis of emilin2a mRNA levels in sorted tcf21:mCherry+ cells (EPDCs) (n=4) and sorted −0.8flt1:RFP+ cells (cECs) (n=4) at 96 hpci normalized to 96 hps. G. RT-qPCR analysis of emilin2a mRNA levels in sorted tcf21:mCherry+ cells (EPDCs) (n=5) normalized to sorted −0.8flt1:RFP+ cells (cECs) (n=5) at 96 hpci. H. Immunostaining of sections of cryoinjured hearts of Tg(flt1:Mmu.Fos-GFP); emilin2a+/+ (H’) and Tg(flt1:Mmu.Fos-GFP); emilin2a−/− (H”) sibling zebrafish at 96 hpci; sections stained for GFP (coronaries, magenta), PCNA (proliferation marker, green), and DNA (DAPI, blue). Arrowheads point to PCNA+ cECs. I. Percentage of PCNA+ cECs in the border zone of Tg(flt1:Mmu.Fos-GFP); emilin2a+/+ (n=5) and Tg(flt1:Mmu.Fos-GFP); emilin2a−/− (n=5) ventricles at 96 hpci. J. Immunostaining of sections of cryoinjured hearts of emilin2a+/+ (J’) and emilin2a−/− (J”) sibling zebrafish at 7 dpci; sections stained for MEF2 (CMs, magenta), PCNA (proliferation marker, green), and DNA (DAPI, blue). Arrowheads point to PCNA+ CMs. K. Percentage of PCNA+ CMs in the border zone of emilin2a+/+ (n=4) and emilin2a−/− (n=4) ventricles at 7 dpci. L. AFOG staining of sections of emilin2a+/+ (L’) and emilin2a−/−(L”) ventricles at 90 dpci. Orange, Muscle; red, Fibrin; blue, Collagen. M. Percentage of scar area relative to ventricular area in emilin2a+/+ (n=4) and emilin2a−/− (n=4) ventricles at 90 dpci. Black (E,L), and white (H,J) dotted lines delineate the injured tissue. Statistical tests: Non-parametric Mann-Whitney test (B,C,D,F,G), Student’s t-test (I,K,M). Scale Bars: 100 μm (E,H,J,L). Ct values of RT-qPCR data are listed in table S3.
