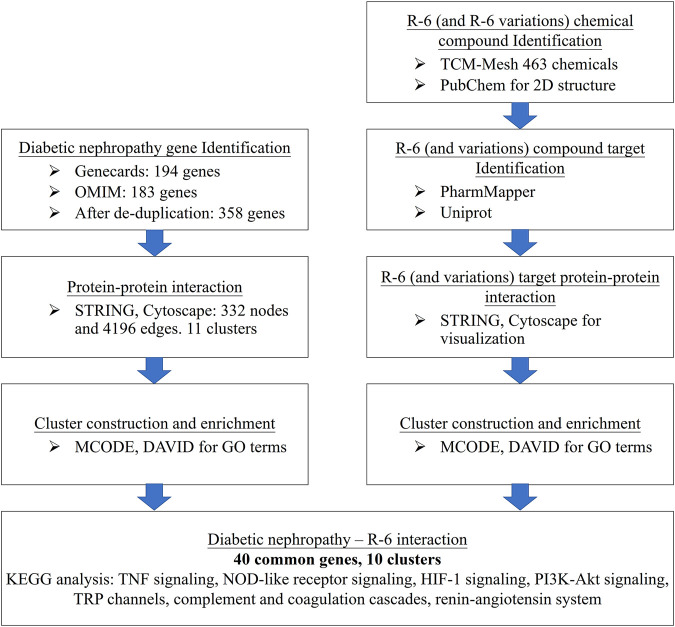
FIGURE 1.
Computational workflow of disease–drug network analysis. Disease-related genes were retrieved from GeneCards and OMIM. The chemical compound IDs of each herb were retrieved from TCM-Mesh and PubChem. Drug targets were revealed via PharmaMapper and UniProtKB. The disease gene interactions were assessed through STRING, and the disease–drug protein–protein interaction network was integrated and visualized by Cytoscape. Functional annotation of clusters was analyzed by DAVID and KEGG pathway enrichment.