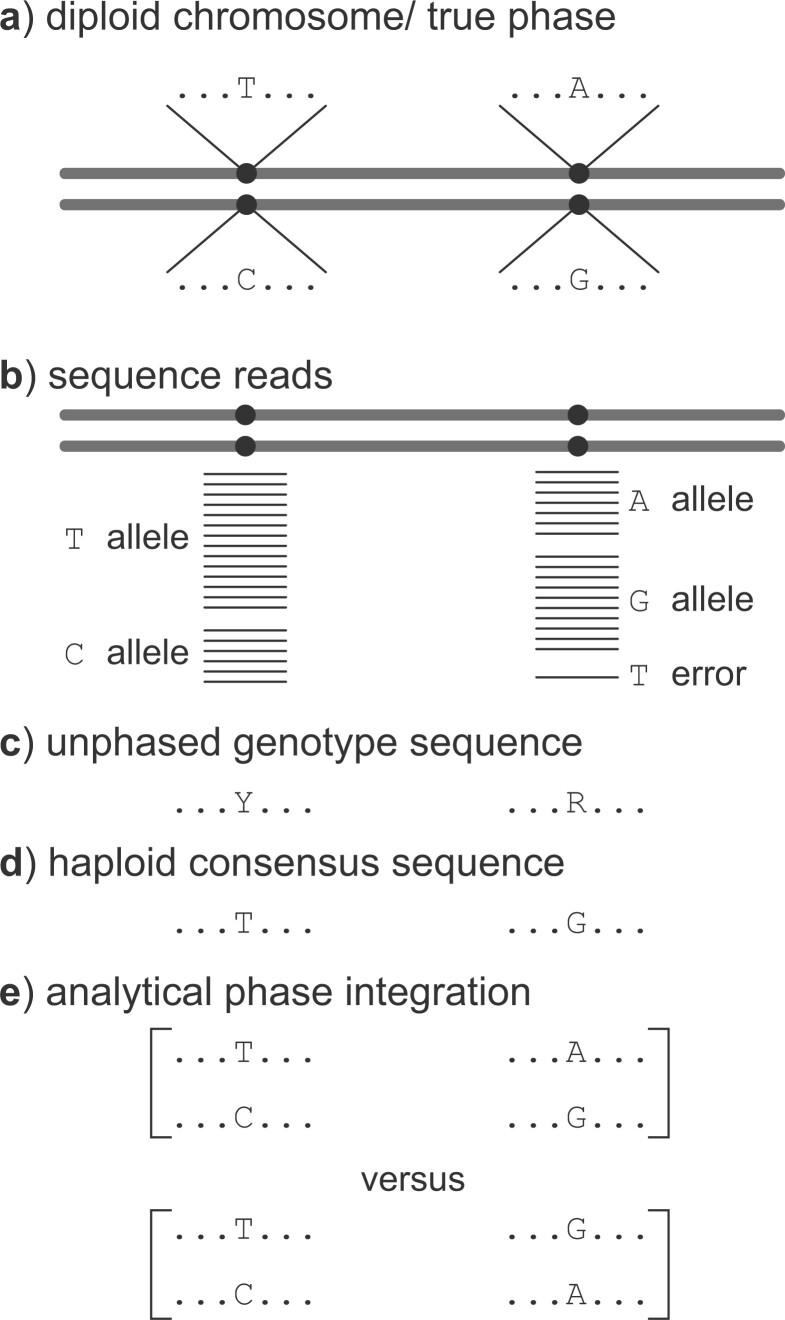
Figure 1.
Example of heterozygote phase resolution. a) A hypothetical diploid chromosome with two heterozygous sites (T/C and A/G). The true haploid genotypes are T...A and C...G. b) Sequence reads around the two heterozygous sites, assuming that they are far apart on the chromosome so that they are not present on any single read (in which case phase would be determined) while they are close enough to be on one locus. In this case, genome assemblers should produce the unphased genotype sequence (c), using the IUPAC ambiguity codes to represent heterozygote sites, but instead they produce the so-called “haploid consensus sequence” (d), picking up the most common nucleotide at each heterozygote site (T...G since T and G are by chance the most common sequence reads at the two sites), which may not match either of the true haploid sequences. e) Analytical integration of phase resolution takes the unphased genotype sequences as data and averages over all possible phase resolutions, weighting each one appropriately according to their relative likelihood based on the whole sequence alignment at the locus.