Figure 5.
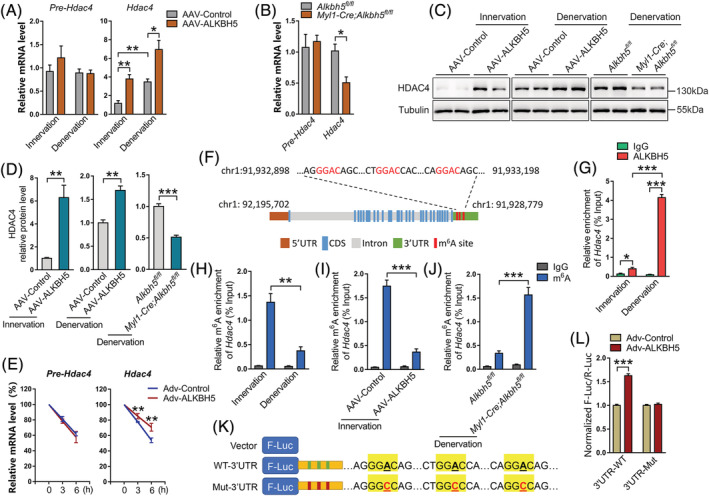
ALKBH5‐mediated m6A demethylation is required for Hdac4 mRNA stability. (A) Precursor and mature mRNA levels of Hdac4 in AAV‐ALKBH5 infected muscle with or without denervation (n = 4). (B) Levels of pre‐Hdac4 and mature Hdac4 in Alkbh5 knockout muscle with denervation (n = 4). (C and D) Protein expression of HDAC4 in ALKBH5 overexpressed and Alkbh5 knockout muscle with or without denervation (n = 4 or 6). (E) Half‐life of pre‐Hdac4 and mature Hdac4 mRNA at 0, 3, and 6 h after actinomycin D (5 μg/mL) treatment in C2C12 myotubes transduced with adenovirus expressing ALKBH5 (Adv‐ALKBH5) (n = 4). (F) Schematic representation of positions of m6A motifs within Hdac4 mRNA. (G) CLIP‐qPCR validation of ALKBH5 binding with 3′UTR of Hdac4 mRNA in both innervated and denervated gastrocnemius (GAS) muscles (n = 4). (H) MeRIP‐qPCR validation of m6A changes in Hdac4 mRNA in muscles during denervation (n = 4). (I and J) m6A‐MeRIP‐qPCR showed that ALKBH5 overexpression depleted m6A modification of Hdac4 mRNA (I), whereas m6A modification of Hdac4 mRNA was enriched after ALKBH5 knockdown (J) (n = 4). (K) Schematic presentation for the generation of wild‐type (WT) or three mutant (Mut; GGAC to GGCC) Hdac4 3′UTR firefly luciferase reporter. (L) Normalized F‐Luc/R‐Luc activity of the WT‐3′UTR and three Mut‐3′UTR reporter in Adv‐ALKBH5 infected C2C12 cells (n = 6). Data are expressed as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 by two‐tailed Student's t test.
