Figure 6.
Species tree estimation error for data simulated from the 16-taxon fungal
tree with a population size of 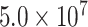 and
varying GDL rates; note that simulations include the effects of GDL and
gene tree estimation error (no ILS). Species tree estimation error was
measured as the normalized RF distance between the true species tree and
the ones inferred from estimated locus trees. The five inference methods
used are ASTRAL,
and
varying GDL rates; note that simulations include the effects of GDL and
gene tree estimation error (no ILS). Species tree estimation error was
measured as the normalized RF distance between the true species tree and
the ones inferred from estimated locus trees. The five inference methods
used are ASTRAL,  ,
NJ
,
NJ , ASTRAL-Pro
(“A-pro”), and FastMulRFS. The duplication/loss rates are
denoted by the rate multiplier (0
, ASTRAL-Pro
(“A-pro”), and FastMulRFS. The duplication/loss rates are
denoted by the rate multiplier (0 ,
1
,
1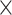 ,
2
,
2 ,
5
,
5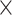 , and
10
, and
10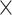 ), where
1
), where
1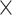 is the rate
estimated in nature for fungi. Each row corresponds to a combination of
population size and GDL rates. The X-axis in each panel represents the
number of gene families used and the Y-axis represents the normalized RF
distance.
is the rate
estimated in nature for fungi. Each row corresponds to a combination of
population size and GDL rates. The X-axis in each panel represents the
number of gene families used and the Y-axis represents the normalized RF
distance.

