Fig. 3.
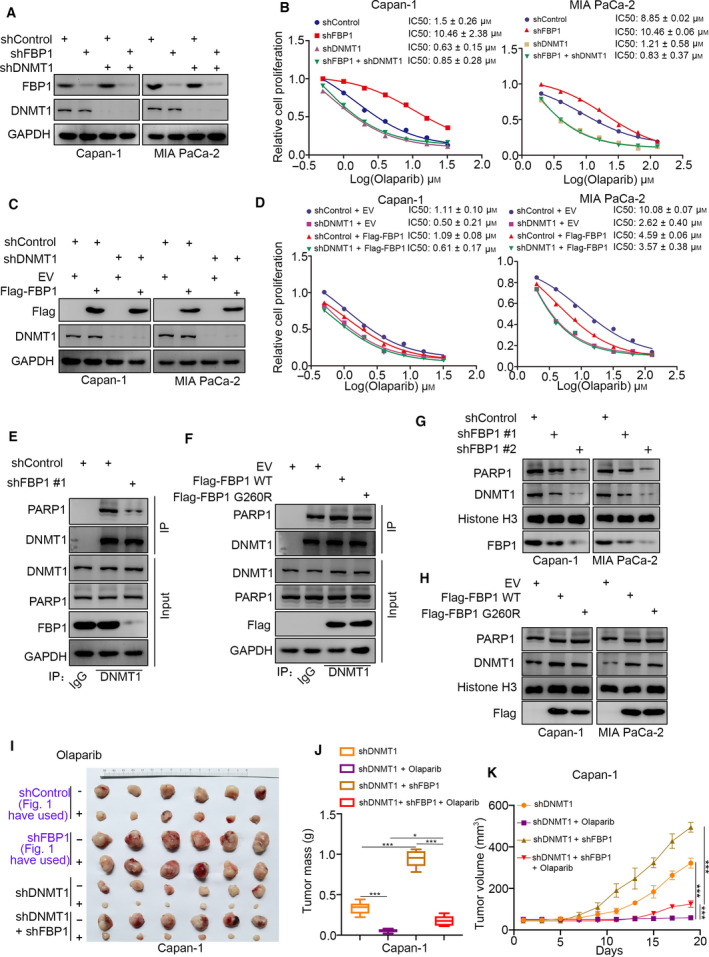
FBP1 determines the sensitivity to DNMT1‐dependent PARP inhibitors. (A and B) MIA PaCa‐2 and Capan‐1 cells were infected with indicated shRNAs for 72 h. Cells were harvested for western blot analysis (A) and treated with a serial concentration of Olaparib for measuring the IC50 values of Olaparib (B), which repeated for three replicates. (C and D) MIA PaCa‐2 and Capan‐1 cells were infected with indicated shRNAs for 48 h. Then, cells were transfected with empty vector or Flag‐FBP1 for another 24 h. Cells were harvested for western blot analysis (C) and treated with a serial concentration of Olaparib for measuring the IC50 values of Olaparib (D), which repeated for three replicates. (E) Capan‐1 cells were infected with indicated shRNAs for 72 h. Cells were harvested and underwent co‐IP assay by using the DNMT1 antibodies, which repeated for three replicates. (F) Capan‐1 cells were transfected with indicated plasmids for 48 h. Cells were harvested and underwent co‐IP assay by using the DNMT1 antibodies, which repeated for three replicates. (G) Capan‐1 cells were infected with indicated shRNAs for 72 h. Cells were harvested and underwent PARP trapping assay, which repeated for three replicates. (H) Capan‐1 cells were transfected with indicated plasmids for 48 h. Cells were harvested and underwent PARP trapping assay, which repeated for three replicates. (I–K) Capan‐1 cells were infected with indicated shRNAs for 72 h. After the puromycin selection, cells were subcutaneously injected into the nude mice. The mice were treated with or without Olaparib (10 mg·kg−1·day−1, i.p., 2 weeks). Representative tumor images (I), tumor weights (J), and tumor growth curves (K) are shown. Data are shown as mean ± SD (n = 6). Statistical analyses were performed with one‐way ANOVA followed by Tukey's multiple comparisons tests. *P < 0.05; ***P < 0.001. The ruler on the top of the representative tumor images on the panel I was used to indicate the specific size of tumors.
