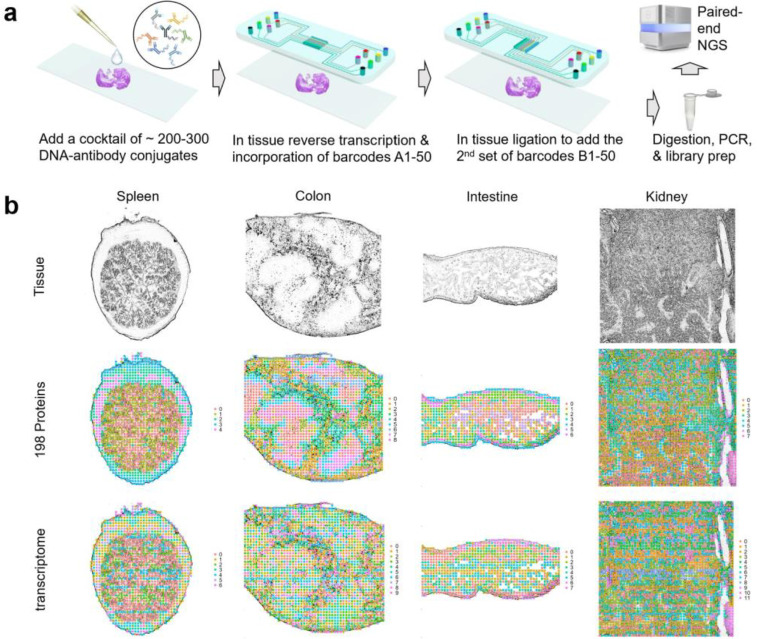
Figure 1. Spatial-CITE-seq workflow design and application to diverse mouse tissue types for co-mapping of 189 proteins and whole transcriptome.
(a) Scheme of spatial-CITE-seq. A cocktail of antibody-derived DNA tags (ADTs) is applied to a PFA-fixed tissue section to label a panel of ~200–300 protein markers in situ. Next, a set of DNA barcodes A1-A50 are flowed over the tissue surface in a spatially defined manner via parallel microchannels and reverse transcription is carried out inside each channel for in-tissue synthesis of cDNAs complementary to endogenous mRNAs and introduced ADTs. Then, a set of DNA barcodes B1-B50 is introduced using another microfluidic device with microchannels perpendicular to the first flow direction and subsequently ligated to barcodes A1-A50, creating a 2D grid of tissue pixels, each of which has a unique spatial address code AB. Finally, barcoded cDNA is collected, purified, amplified, and prepared for paired end NGS sequencing. (b) Spatially resolved 189-plex protein and whole transcriptome co-mapping of mouse spleen, colon, intestine, and kidney tissue with 20μm pixel size. Upper row: brightfield optical images of the tissue sections. Middle row: unsupervised clustering of all pixels based on all 189 protein markers only and projection onto the tissue images. Lower row: unsupervised clustering of whole transcriptome of all pixels and projection to the tissue images. Colors correspond to different proteomic or transcriptomic clusters indicated on the right side of each panel.