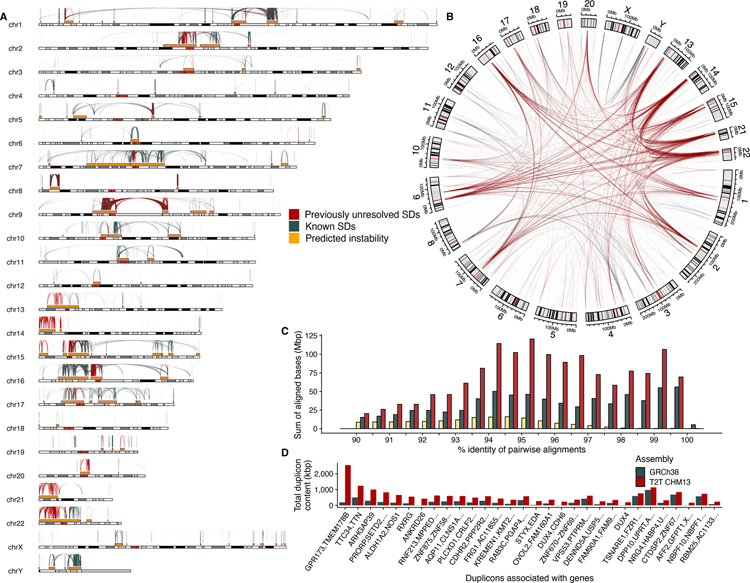
Fig. 1. Segmental duplication (SD) content of the T2T-CHM13 genome.
A) The pattern of previously unresolved or structurally variant intrachromosomal duplications in T2T-CHM13 (red) compared to known duplications in GRCh38 (blue-gray). These predict hotspots of genomic instability (gold) flanked by large (>10 kbp), high-identity (>95%) interspersed (>50 kbp) SDs. B) Circos plot highlighting previously unresolved interchromosomal SDs (red) shows the preponderance of previously unresolved SDs mapping to pericentromeric and acrocentric regions. C) A histogram comparing SD content in different human reference genomes. The sum of bases in pairwise SD alignments stratified by their percent identity for the Celera (yellow, Sanger-based), GRCh38 (blue-gray, BAC-based), and T2T-CHM13 (red, long-read) assemblies. D) The 30 genic duplicons (ancestral repeat units) with the greatest copy number difference between GRCh38 and T2T-CHM13 as determined by DupMasker (table S2). All of the 30 largest differences are increased in T2T-CHM13.