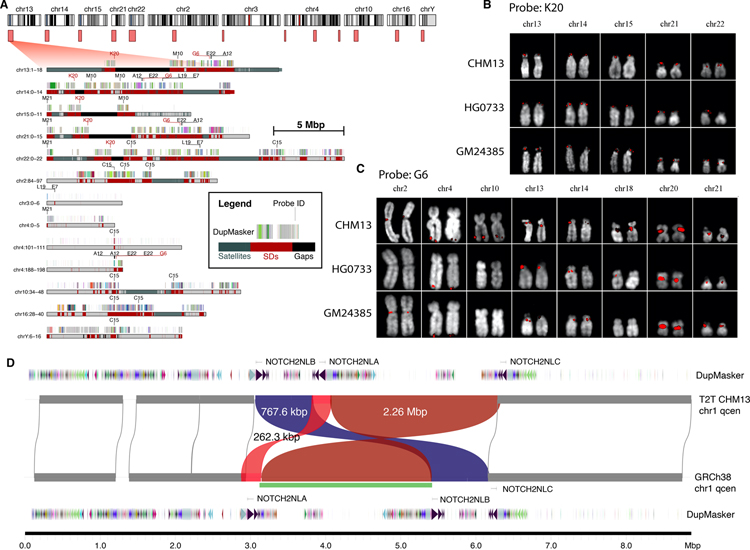
Fig. 2. Validation of previously unresolved SDs in T2T-CHM13 and heteromorphic variation.
A) Ideogram (top) depicts large SD regions (light red boxes) present in T2T-CHM13 but absent from the current reference human genome (GRCh38). An expanded view of the duplication (red) and satellite organization (blue) are depicted below showing the location of fosmid FISH probes (e.g., C15) and SD organization compared to ancestral duplicon segments (multi-colored bars) (see inset). B,C) FISH signals (red) shown on extracted metaphase for two probes and three human cell lines. Probe K20 shows a fixed signal (except for one heterozygous signal), and G6 is heteromorphic among humans (see table S4 and fig. S4 for complete description for all nine probes). D) Inversion polymorphism (green bar) between T2T-CHM13 and GRCh38 in the pericentromeric chromosome 1q region. The inversion (green bar) identified by Strand-seq (32) is confirmed in the assembly; however, the sequence-resolved assembly shows a more complex structure including two inversions (red) and one reordered segment (blue) mapping near the NOTCH2NL human-specific duplications.