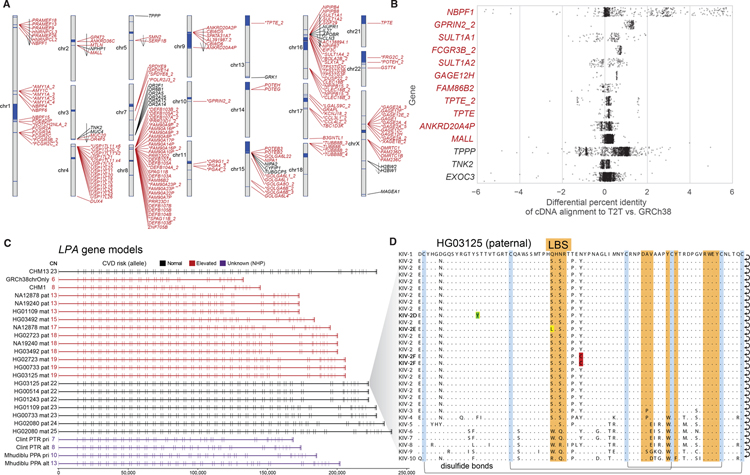
Fig. 5. Genic variation in previously unresolved SD regions of T2T-CHM13.
A) Ideogram showing the previously unresolved or non-syntenic gene models (open reading frames [ORFs] with >200 bp of coding sequence and multiple exons) in the T2T-CHM13 assembly as predicted by Liftoff. Previously unresolved genes mapping to SDs (red) are indicated with an asterisk if predicted to be an expansion in the gene family relative to GRCh38 (25). Arrows indicate inverted regions. Most unique genes mapping to non-syntenic regions (black) are the result of an inversion (arrow). B) Percent improvement in mapping of CHM13 Iso-Seq reads in candidate duplicated genes (red) mapping to non-syntenic regions of the T2T-CHM13 assembly. Positive values identify Iso-Seq reads aligning better to T2T-CHM13 than GRCh38. C) Gene models of LPA with ORF generated from haplotype-resolved HiFi assemblies. The double-exon repeat in these gene models encode for the Kringle IV subtype 2 domain of the LPA protein. Highlighted in red are haplotypes with reduced Kringle IV subtype 2 repeats predicted to increase risk of cardiovascular disease. D) Amino acid variation in the Kringle IV subtype 2 repeat in the paternal haplotype of HG01325 identifies a previously unknown set of amino acid substitutions including rare variants: Ser42Leu in the active site, Ser24Tyr and Tyr49Cys.