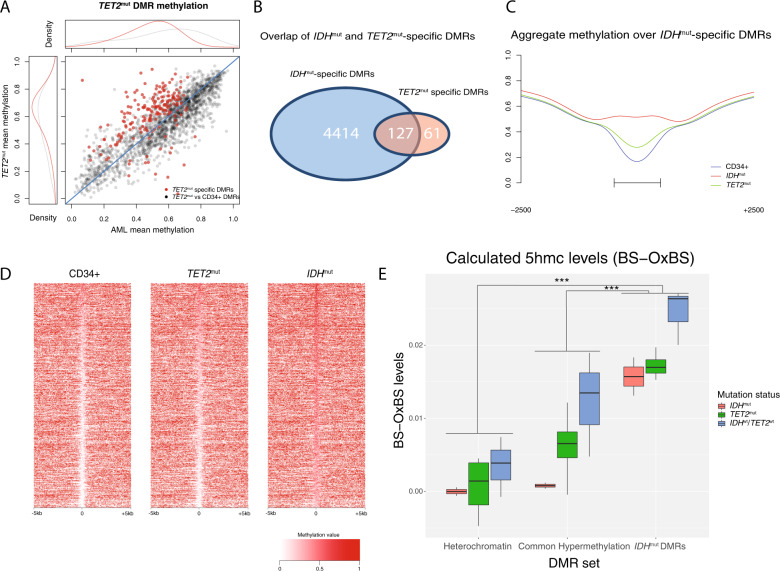
Fig. 3. TET2mut AMLs have modest hypermethylation that overlaps IDHmut-specific DMRs.
A Mean DMR methylation across 1879 TET2mut DMRs called vs. CD34+ cells (black points) and 188 in TET2mut-specific DMRs (red points) in TET2 mutant samples versus all other AMLs that are wild type for IDH, TET2, and DNMT3A. B Intersection of TET2mut-specific and IDHmut-specific DMRs. C Aggregate methylation over IDHmut-specific DMRs in IDHmut and in TET2mut AML and CD34+ cells. D Locus heatmap of mean methylation values for all IDHmut-specific DMRs (rows), where each column is centered over the DMR with the window extending 5 kb up- and downstream the DMR center point. E Mean 5hmC (WGBS minus oxWGBS) levels in IDHmut, TET2mut, and IDHwt/TET2wt AML samples at 105,519 ChromHMM heterochromatic regions, 4586 generically hypermethylated regions (i.e., regions that were hypermethylated vs. CD34+ cells in at least two AMLs without IDH1, IDH2, or TET2 mutations), and 4541 IDHmut-specific hypermethylated DMRs.