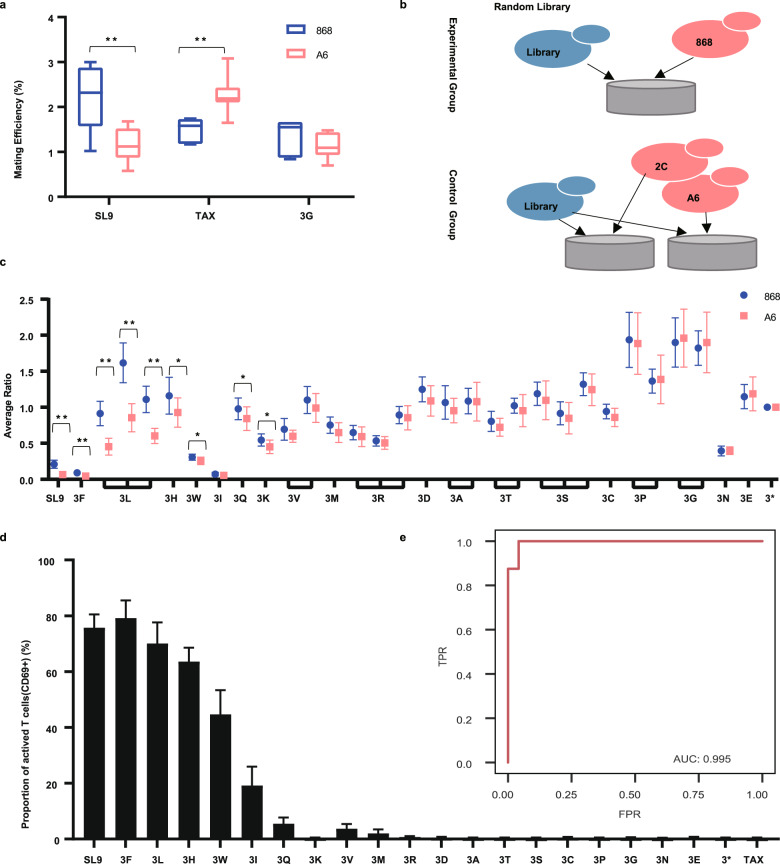
Fig. 3. The YAMTAD system can be used to screen cognate peptides for a given TCR.
a The mating efficiency (mean ± sd) between the cognate TCR-pMHC pairs was significantly higher than that of the negative control pairs. The mating efficiency was calculated by the percentage of diploids detected by FACS. Seven biological repeats were measured. b A schematic diagram of the strategy used for screening cognate peptides for a given TCR in a randomly generated peptide library. The random peptide library was mated with MATa yeasts expressing the 868 TCR as the experimental group and with MATa yeasts expressing the A6 or 2 C TCR as the control group. c Ratios (mean ± sd) of the indicated SL9 mutants to the 3* peptide in diploid yeasts induced in YAMTAD screening. Each experiment was performed with 4 technical repeats and was independently duplicated 3 times. The mean of 12 duplicates is shown. d Proportion (mean ± sd) of 868 T cells activated by various SL9 mutants in the yeast T cell co-culture system. The Y-axis represents the proportion of activated T cells (CD69+) after coculturing with the indicated yeast strains. e The ROC curve depicting the FPR (false-positive rate, 1-specificity) and TPR (true positive rate, sensitivity) for library screening of TCR-pMHC interactions using the YAMTAD system. We defined a peptide as a cognate peptide if it could activate more than 10% of the T cells on average in the yeast T cell coculture assay. Unpaired two-tailed Student’s t-test was used for statistical analysis in a, c. **Indicates a two-tailed P value < 0.01. *Indicates a two-tailed P value < 0.05.