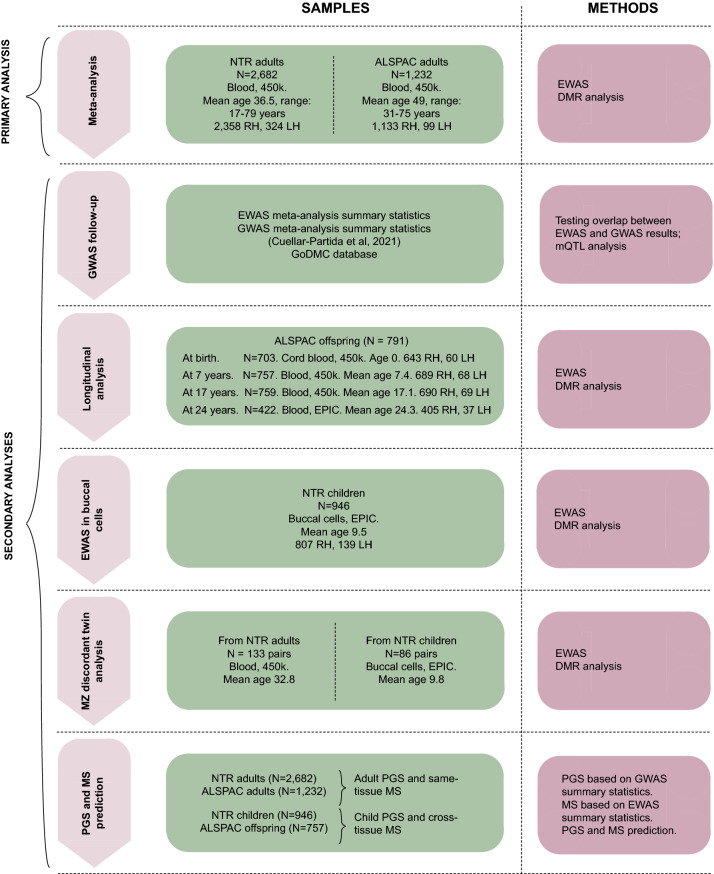
Figure 1.
Flowchart of epigenome-wide association study of left-handedness. The flowchart summarizes the study design. The primary analyses included EWASs in NTR adults and ALSPAC adults, followed by meta-analysis to identify DNA methylation sites associated with left-handedness. The secondary analyses included: (1) left-handedness GWAS loci follow-up; (2) longitudinal analysis of DNA methylation at four ages in ALSPAC offspring; (3) analysis of buccal cell DNA methylation in NTR children; (4) analysis of DNA methylation differences between left and right-handed co-twins from NTR discordant MZ twin pairs and (5) polygenic and DNA methylation scores prediction. For left-handedness prediction, polygenic scores (PGS) were created based on left-handedness GWAS summary statistics not including NTR/ALSPAC. Methylation scores (MS) were created based on weights from EWASs in NTR adults, ALSPAC adults, NTR children and ALSPAC offspring at 7 years old to estimate the predictive performance. LH, left-handed; RH, right-handed. DMR, differentially methylated region. EWAS, epigenome-wide association study. GWAS, genome-wide association study. GoDMC, Genetics of DNA Methylation Consortium. mQTL, methylation quantitative trait locus. Blood, buccal cells indicate tissue of DNA methylation. 450 k, EPIC indicate the platform for DNA methylation measurement.