Abstract
Lichens are used in folklore medicines across the globe for wound healing and to treat skin disorders and respiratory diseases. They are an intricate symbiosis between fungi and algae with the domination of fungal counterparts. Recent research studies pointed out that yeast is a third major partner in lichens. Endolichenic fungi (ELF) are also a part of this complex miniature ecosystem. The highly competitive environment of lichens compels ELF to produce toxic metabolites which are comparatively less explored for their chemical diversity and use. Here, we investigated 31 ELF isolated from 32 lichens found on mangrove plants at Puttalam Lagoon of Sri Lanka to find cytotoxic molecules by applying LC-UV-HRMS analysis and in vitro bioassays. The studies resulted in the identification of three potent cytotoxic molecules from endolichenic fungi Talaromyces pinophilus isolated from host lichen Porina tetracerae. The ethyl acetate extract of this fungus showed moderate cytotoxicity against the breast cancer cell line. Chemical characterization of ethyl acetate extract of T. pinophilus produced peniazaphilin B, 152G256α-1, and ES-242-3. The structures of these molecules were confirmed by NMR and MS data. We are reporting ES-242-3 for the first time from the genus Talaromyces and peniazaphilin B and 152G256α-1 from T. pinophilus. The isolated compounds were evaluated for their anticancer potential against breast, oral and cervical cancer cell lines. Compound 152G256α-1 showed potent cytotoxicity against oral cancer (CAL-27 cell line) with an IC50 value of 2.96 ± 0.17 µM while ES-242-3 showed the best activity against breast cancer (MCF-7 cell line) and cervical cancer (HeLa cell line) with IC50 value 14.08 ± 0.2 µM and 4.46 ± 0.05 µM respectively. An in-silico analysis was carried out to predict the mechanism of in-vitro activity, drug likeliness, and pharmacokinetic profile of the isolated compounds. The study confirms the potential of ELF T. pinophilus to produce diverse bioactive scaffolds and encourages the researchers to further explore the fungus and its metabolites with newer technologies to produce potent anticancer leads.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s12088-021-00994-8.
Keywords: Talaromyces pinophilus, Peniazaphilin B, 152G256α-1, ES-242-3, Cytotoxic, Anticancer
Introduction
Drug resistance in cancer therapy is a multifaceted challenge due to numerous factors like tumor microenvironment, tumor heterogeneity, immune system, tumor burden, growth kinetics, etc. that contribute to its complexity. Scientists have proposed multiple solutions to tackle this issue, major being the discovery of novel drugs [1, 2]. Natural Products is a well-established source of unique scaffolds having potent pharmacological activities. In cancer therapy, above 60% of drugs are based on Natural Products such as podophyllotoxin, camptothecin, paclitaxel, and vinblastine [3, 4]. The landscape of bioactive compounds from nature indicate that one of the major contributor is fungi underlining its significant role in the process of drug discovery [5]. Lichen is one of the unique biodiverse ecosystems consisting of fungi, algae and other microorganisms. It holds an important place in folk medicine and is a source of thousands of secondary metabolites [6]. However, the origin of these bioactive metabolites is still not well defined, it could either be lichen-forming fungi or other associated microbes. Lichens are slow-growing and difficult to harvest in larger quantities and therefore, researchers are trying to find culturable microorganisms that are obtained from lichen and are able to produce bioactive secondary metabolites. Endolichenic fungi (ELF) are one of such organisms residing within the lichen thalli and only 2% of the total diversity of ELF has been explored in terms of its chemical potential [7]. ELF residing within mangrove based lichens can produce unique secondary metabolites as they are exposed to the harsh environment due to high humidity and salinity. Considering this rationale, the current study was planned to explore the chemical diversity of endolichenic fungi isolated from lichens residing on mangrove plants of Puttalam lagoons from Sri Lanka.
A study of 32 lichens from Puttalam Lagoon, Sri Lanka resulted in 171 ELF belonging to various genera like Aspergillus, Byssochlamys, Talaromyces, Daldinia, Diaporthe, Phomopsis, Cerrena, Xylaria, Endomelanconiopsis, Schizophyllum, Trichoderma, Hypoxylon, Preussia, Sordaria, Lasiodiplodia, and Neurospora [8]. Here, we have screened ethyl acetate extracts of 31 ELF from the above-mentioned fungus species using LC-HRMS based cluster analysis [9] and investigated Talaromyces pinophilus for its bioactive secondary metabolites. The noteworthy potential of fungi from genus Talaromyces to produce diverse chemical scaffolds has been evident from the published reports [10]. Three bioactive compounds were isolated from T. pinophilus and evaluated their cytotoxicity against breast cancer (MCF-7), cervical cancer (HeLa), and oral cancer (CAL-27) cell lines. In addition, drug likeliness, pharmacokinetic properties, and its interaction with various receptors associated with the respective cell lines were predicted through in-silico studies.
Materials and Methods
General Methods
Jasco binary HPLC system with ChromPass software was used for the analysis of extracts, fractions, and pure compounds. Phenomenex C18, 250 × 4.60 mm (L × I.D.), 5 µm particle size column was used. The mobile phase was acetonitrile and 0.1% formic acid in DI water with gradient elution. Agilent 1290 Infinity UHPLC system connected to QTOF 6545 instrument was used for LC-HRMS analysis of the samples. Poroshell 120 SB C18, 2.7 µm particle size, L × I.D. 4.6 × 100 mm column was used. Gradient elution at 0.4 mL/min flow rate with initial hold up to 3 min at 10% methanol in 0.1% v/v of formic acid in DI water, which was increased to 95% from 3 to 9 min and held at 95% till 11 min. From 11 to 15 min the solvent composition adjusted to starting condition for the equilibration and next injection readiness. PDA (254 nm) and Q-TOF (100–1000 Da) detectors were used for the analysis. The data was analysed using Masshunter software. Agilent 1260 infinity semi-preparative HPLC system, Zorbax RX C18, 250 × 9.4 mm, 5 µm particle size column were used for purification of compounds. Mobile phase initial hold of 5 min was kept at 5% acetonitrile and 95% of DI water having 0.1% formic acid, the gradient increase of solvent was given from 5 to 95% in next 10 min followed by a hold at 95% from 15 to 25 min. Bruker 8150 FTIR instrument was used to acquire FTIR spectrum in the range of 400–4000 cm−1 and analyzed using OPUS 7.5 software (Bruker, Germany). Thermo Scientific Evolution 300 instrument was used for UV specific absorptivity analysis. Jasco J815 model was used for CD analysis which was performed at central instrument facility, Indian Institute of Technology (IIT), Gandhinagar. The NMR spectra 1D, and 2D were acquired on Bruker’s 500 MHz instrument (Bruker, Avance NEO, MA, USA) using solvents like CDCl3 and MeOD.
Culturing and Extraction of T. pinophilus
Isolation of ELF from lichens belonging to mangrove plants of Puttalam Lagoon, Sri Lanka has been discussed in the earlier article [8]. T. pinophilus isolated from the host lichen Porina tetracerae was cultured on potatoes dextrose agar (100 plates; plate size 10 × 20 mm) incubated for 14 days. The agar media was prepared by using 210 g of potatoes, 168 g of glucose, and 161 g of Agar for 7 L of media. The mycelia along with the media were cut into small pieces and extracted with ethyl acetate (6 × 500 mL) followed by evaporation of the solvent under reduced pressure. The extract was transferred to glass vials and purged with N2 to remove the residual solvent and stored at 0 °C [8].
Isolation and Characterization of Compounds
Ethyl acetate extract of T. pinophilus culture (820 mg) was subjected to normal phase (neutral silica #230-400) chromatography with a step gradient of hexanes: ethyl acetate combinations (100% hexanes to 100% ethyl acetate; step of 10%). The fractions eluting from the column were collected and monitored using TLC and HPLC and similar profile fractions were mixed leading to 21 fractions. The semi-preparative HPLC method was utilised for the purification of enriched fractions 11 and 12. Zorbax SB-C18, 9.4 × 250 mm, 5 µm particle size column was used. Gradient elution method was used with acetonitrile and 0.1% v/v formic acid in DI water. Fraction 11 led to the isolation of peniazaphilin A (4.5 mg), and 152G256α-1 (3.1 mg), while compound ES-242-3 (2.4 mg) was isolated from fraction 12.
Characterisation of the Isolated Compounds
Peniazaphilin B
HRMS m/z: 209.0827 [M + H]+; Molecular formula: C11H12O4; FTIR νmax cm−1: 3550(OH), 1740 (C = O); UV (MeOH) (λmax) ε: 214 (4.3), 263.5 (4.1) and 298 (3.7) nm; CD (MeOH) Δ ε (nm): − 7.37 (231), + 1.38 (250), − 6.69 (269); 1H NMR (500 MHz, CDCl3) (δ) 4.54 (H-2,m), 2.82 (H-3, m), 6.33 (H-4, s), 6.43 (H-6, s), 3.82 (H-10, s), 1.46 (H-11,d, J = 5 Hz); 13C NMR (125 MHz, CDCl3) (δ) 163.82 (C-1), 73.88 (C-2), 36.20 (C-3), 106.5 (C-4), 162.8 (C-5), 98.69 (C-6), 165.53(C-7), 105.75 (C-8), 144.0 (C-9), 55.95 (C-10), 220.6 (C-11).
152G256α-1
HRMS m/z: 663.2298 [M + H]+; Molecular formula:: C32H38O15; FTIR νmax cm−1: 3500 (OH), 2935 (C-H), 1736 (C = O), 1052 (C-O); UV (MeOH) (λmax) ε: 4.6 (217), 4.2 (265) and 3.9 (303) nm; CD (MeOH) Δ ε (nm): − 18.79 (212), + 5.92 (231), + 10.23 (268) 1H NMR (500 MHz, CDCl3) (δ) 6.32 (H-8, d, J = 2.4 Hz), 6.29 (H-24, d, J = 2.4 Hz), 6.24 (H-22, d, J = 2.4 Hz), 6.14 (H-10, d, J = 2.1 Hz), 1.49 (H-18, d, J = 6.3 Hz), 1.44 (H-4, d, J = 6.3 Hz), 1.29 (H-28, d, J = 6.2 Hz), 1.18 (H-14, d, J = 6.2 Hz), 5.66 (H-17,m), 5.52 (H-3, m), 5.22 (H-27, m), 5.15 (H-13, m), 4.32 (H-31), 4.07 (H-32,m), 3.51 (H-12a,m),2.89 (H-12b, m), 3.17 (H-26a, d, J = 6.8 Hz) 2.86 (H-26b, m), 2.84 (H-16a, m), 2.81 (H-16b, m), 2.76 (H-2a, m), 2.70 (H-2b, m), 2.52 (H-30a, m), 2.46 (H-30b, m); 13C NMR (125 MHz, CDCl3) (δ) 171.37 (C-1), 40.48 (C-2), 69.22 (C-3), 20.11 (C-4), 171.34 (C-5), 105.66 (C-6), 165.03 (C-7), 102.39 (C-8), 160.86 (C-9), 111.60 (C-10), 142.33 (C-11), 40.74 (C-12), 71.74 (C-13), 19.78 (C-14), 170.24 (C-15), 41.07 (C-16), 69.03 (C-17), 19.80 (C-18), 170.10 (C-19), 105.15 (C-20), 165.36 (C-21), 102.40 (C-22), 160.97 (C-23), 110.84 (C-24), 142.08 (C-25), 40.66 (C-26), 72.20 (C-27), 19.42 (C-28), 170.40 (C-29), 38.37 (C-30), 66.30 (C-31), 67.32 (C-32).
ES-242-3
HRMS m/z: 621.2331 [M + H]+; Molecular formula: C34H36O11; FTIR νmax cm−1: 3392(OH),2937 (C-H), 1733 (C = O), 1097 (C-O); UV (MeOH) (λmax) ε: 4.9 (239), 4.1 (296), 4.1 (308), 4.0 (339) and 4.1 (354) nm; CD (MeOH) Δ ε (nm): + 1.84 (239), − 4.16 (252), − 0.24 (339) 1H NMR (500 MHz, MeOD) (δ) 6.58 (7-H, d, J = 2.3 Hz), 6.55 (H-7’, d, J = 2.2 Hz), 6.10 (H-5, d, J = 2.3 Hz), 5.85 (H-5’d, J = 2.2 Hz), 5.31 (H-4, d, J = 1.6 Hz, 1H), 5.17 (H-1a, d, J = 15 Hz), 5.20 (H-1’a, d, J = 15 Hz), 4.78 (H-1b, d, J = 15 Hz), 4.80 (H-1b’, d, J = 15 Hz),3.88 (H-4’, d, J = 1.7 Hz), 3.86 (H-3’, m), 3.76 (H-3, m), 1.18 (H-11, d, J = 6.4 Hz), 1.01 (H-11’, d, J = 6.4 Hz), 3.41 (H-6-OCH3, s), 4.07 (H-8-OCH3, s), 1.06 (H-4-O(C = O)CH3, s), 3.42 (H-6’-OCH3, s), 4.06 (H-8’-OCH3, s) 13C NMR (125 MHz, MeOD) (δ) 64.62 (C-1), 73.93 (C-3), 66.07 (C-4), 130.01 (C-4a), 98.12 (C-5), 157.49 (C-6), 98.13 (C-7), 157.17 (C-8), 110.28 (C-8a), 149.53 (C-9), 114.34 (C-9a), 125.89 (C-10), 135.49 (C-10a), 15.90 (C-11), 54.14 (6-OCH3), 54.09 (8-OCH3), 169.81 (4-O(C = O), 17.93 (4-O-(C = 0)CH3)64.50 (C-1’), 73.31 (C-3’), 67.09 (C-4’), 135 (C-4’a), 99.14 (C-5’), 157.10 (C-6’), 97.35 (C-7’), 157.42 (C-8’), 110.04 (C-8’a), 149.39 (C-9’), 114.18 (C-9’a), 123.92 (C-10’), 136.10 (C-10’a), 15.80 (C-11’), 55.66 (6’-OCH3), 55.68 (8’-OCH3).
In-Vitro Anticancer Activity
Anticancer activity of extracts were tested against MCF-7 cell line using Alamar Blue assay [11] in the concentration range of 5, 50, 100, and 150 µg/mL. The pure compounds were evaluated for their anticancer activity against MCF-7 (Breast cancer) cell line at the concentration of 1, 10, 50, and 100 µM and, HeLa (cervical cancer) and CAL-27 (Oral cancer) cell lines at the concentration of 1, 5, 10 and 20 µM. The compounds were evaluated for cytotoxicity on normal human breast cell line MCF-10A at concentrations 5, 10, 50, and 100 µM. The fluorescence was measured using the Varioscan Go instrument with Scanit software. The data were analysed using Graphpad prism software.
In-Silico Studies
Protein Structure Selection and Retrieval
Human estrogen receptor (ESR1) and WNT-7B receptors were selected as target proteins for MCF-7 cell lines to represent Breast cancer. The structural and functional details of ESR1 and WNT-7B were obtained from Universal Protein Resource (UniProtKB) database with UniProt ID P03372 and P56706 respectively. The experimental structure of ESR1 was retrieved from RCSB (Research Collaboratory for Structural Bioinformatics), PDB (Protein Data Bank) with ID 3ERT, having resolution 1.90 Å. As the experimental structure of protein WNT, − 7B was not available in PDB, the structure was predicted through homology modelling using modeller 9.21. The amino acid sequence of the protein was retrieved from UniProt ID P56706 which was further analysed using BLAST for getting templates (4F0A-B chain and 6AHY-B chain). Subsequently, programmed cell death 6-interacting protein (ALIX) and tumor susceptibility gene 101 protein (TSG101) were selected for oral cancer while their structural and functional details were obtained from the UniProtKB database with UniProt ID Q8WUM4 and Q99816 respectively. The experimental 3D structures of ALIX and TSG101 were obtained from RCSB, PDB with PDB ID 3C3R (Resolution: 2.02 Å) and 3OBQ (Resolution: 1.40 Å). In the case of cervical cancer, transferrin receptor protein (TFRC) was selected with UniProt ID P02786 and structure was obtained from RCSB, PDB with ID 1DE4 having resolution 2.80 Å [12, 13].
Prediction of Binding Pockets
The binding site for ESR1 was predicted through PDBsum online database whereas for WNT7B, ALIX, TSG101 and TFRC was predicted through CASTp, DEPTH and Ghecom online tools [14].
Molecular Docking
The structures of pure compounds were docked against the selected abovementioned proteins using AutoDock 4.2. To efficiently dock with ligands/compounds the grids were generated for each of the proteins. The space dimension parameters for them were a) ESR1: x-centering 72, y-centering 58 and z-centering 66; b) WNT7B: x-centering 126, y-centering 126 and z-centering 126; c) ALIX: x-centering 120, y-centering 90 and z-centering 126; d) TSG101: x-centering 62, y-centering 74 and z- centering 80; e)TFRC: x-centering 124, y-centering 104 and z-centering 94. Consequent docking complexes were evaluated for binding energy scores, ligand efficiency, intermolecular H-bonds and other interactions. The interactions are visualised in Discovery Studio Visualizer [15].
Drug Likeliness and Pharmacokinetics
Molinspiration, Molsoft, ADMET SAR, and Swiss ADME were used to assess the drug likeliness of molecules according to the number of hydrogen bond acceptors (HBA) and hydrogen bond donors (HBD), molecular mass, and Log P-value. Furthermore, their predicted absorption, distribution, metabolism, and excretion (ADME) properties were evaluated by Pre ADMET and the Swiss ADME online tool [16].
Results and Discussion
Cluster analysis is a technique of grouping similar sets in one cluster. This helps in analyzing the patterns, similarities, and differences in the data set [9]. In this study, the LC-UV-HRMS data of ethyl acetate extracts of 31 endolichenic fungal strains were analysed thoroughly for the prominent peaks and their patterns for segregation into three clusters Priority 1, Priority 2, and Priority 3. The prioritising of extracts was based on UPLC-UV (254 nm) as well as Mass profile. Here, the extracts with a single major peak or 2–3 major peaks were placed in the priority 1 group. The extracts showing the presence of 5–7 moderate height peaks were placed in the priority 2 group and lastly, the extracts with less intense peaks were placed in priority 3. Ethyl acetate extracts of Talaromyces pinophilus, Lasiodiplodia crassispora, Byssochlamys spectabilis, and Endomelanconiopsis sp. have qualified for priority 1 (Table 1). Anti-cancer activity of these priority 1 extracts was evaluated in vitro against human breast cancer cell line MCF-7. The observed IC50 value of all the screened extracts after treatment of 24 h have been depicted in Table 2. Ethyl acetate extract of T. pinophilus showed the best IC50 value 37.03 µg/mL among all tested extracts and hence, the fermentation of this culture at higher volume and further investigated to isolate bioactive compounds (Fig. 1).
Table 1.
Clusters of ELF species according to LC–UV–MS analysis
| Priority 1 | Priority 2 | Priority 3 |
|---|---|---|
| Talaromyces pinophilus | Diaporthe arengae | Nigrospora sp. |
| Lasiodiplodia crassispora | Xylariaceae sp. | Phomopsis sp. |
| Byssochlamys spectabilis | Nigrospora sphaerica | Hypoxylon anthochroum |
| Endomelanconiopsis sp. | Lasiodiplodia theobromae | Neurospora sp. |
| Aspergillus hiratsukae | Xylaria psidii | |
| Xylariaceae sp. | Xylaria feejeensis | |
| Neosartorya hiratsukae | Daldinia eschscholtzii | |
| Chaetomium fuscum | Lasiodiplodia pseudotheobromae | |
| Preussia tenerife | Hypoxylon anthochroum | |
| Xylaria castorea | Nodulisporium sp. | |
| Neurospora crassa | Rigidoporus vinctus | |
| Aspergillus aculeatus | Aspergillus fumigatus | |
| Endomelanconiopsis sp. | Schizophyllum commune |
Table 2.
In-vitro cytotoxicity assay data of priority 1 extracts
| Sr. no. | Name of Fungi (Ethyl acetate extract) | IC50 against MCF-7 |
|---|---|---|
| 1 | Talaromyces pinophilus | 37.03 ± 0.7 μg/mL |
| 2 | Lasiodiplodia crassispora | 75.97 ± 0.3 μg/mL |
| 3 | Byssochlamys spectabilis | 96.23 ± 0.4 μg/mL |
| 4 | Endomelanconiopsis sp. | 88.45 ± 0.6 μg/mL |
| 5 | Doxorubicin (Positive control) | 1.31 ± 0.06 μM |
Fig. 1.
Culture of endolichenic fungi Talaromyces pinophilus
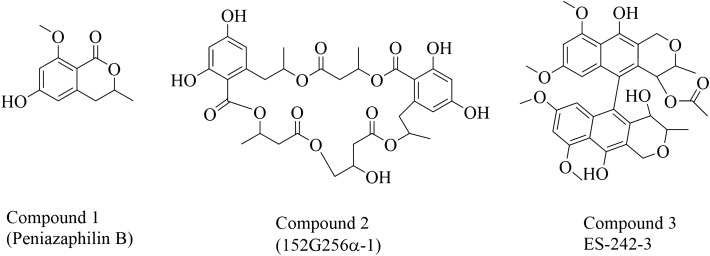
Normal phase (Si) column chromatographic separation followed by reversed-phase semi-preparative HPLC resulted in three pure compounds. Characterisation of the isolated compounds was done using UV, FTIR, HRMS, CD, and NMR (refer to supplementary information). Compound 1 was obtained as a pale yellow powder. The empirical formula was deduced as C11H12O4 with 6 degrees of unsaturation according to the observed [M + H]+ 209.0805 m/z, [M + Na]+ 231.0618 m/z. It was further confirmed by 1H, 13C and DEPT-135 NMR data. A literature search of molecular formula, accurate mass and NMR data in Dictionary of Natural Products (DNP) have revealed that the compound could be peniazaphilin B, which is reported from Penicillium sp. CPCC 400,786 [17] and Talaromyces thailandiasis [18], although no reports were available from species T. pinophilus. The careful comparison of 1H, 13C, DEPT, COSY, HSQC and HMBC data of compound 1 with the reported data of peniazaphilin B confirmed its 2D structure (Fig. 2a). Circular dichroism data confirmed its 3R configuration evident by negative cotton effect at 231 and 269 nm and positive cotton effect at 250 nm [17].
Fig. 2.
Structures of isolated compounds from endolichenic fungi Talaromyces pinophilus
The compound 2 was isolated from fraction 11 as a white powder with [M + H]+ at 663.2298 m/z, empirical formula C32H38O15 (14 degrees of unsaturation). 1H and 13C and DEPT-135 data of 2 indicated the presence of 4 methyl, 6 methylene, 5 methine, and 13 quaternary carbons. The 13C NMR signals at δ 171.37, 171.34, 170.24, 170.10, and 170.40 indicated 5 carbonyls and 1H chemical shifts at δ 5.52, 5.13, 5.66, 5.22, and 4.30 with corresponding carbon chemical shifts at δ 69.22, 71.13, 69.03, 72.20, and 66.30 revealing macrocyclic pentalactone scaffold. This partial structure was searched in DNP and Scifinder with filters of molecular formula and biological source (genus) showed the probability of 152G256α, 152G256α-1 and 152G256o [19]. Further, long-range HMBC correlation showcasing the γ-hydroxy ester linkage narrowed the hits to 152G256α-1. This compound was earlier reported from Talaromyces flavus [19] and for the first time, we are reporting from T. pinophilus. The CD spectra comparison confirmed its 3D structure (Fig. 2b).
LC-HRMS analysis of compound 3, isolated as a pale yellow powder, showed [M + H]+ 621.2331 m/z, [M + Na]+ 643.2149 m/z, and [M + K]+ 659.1891 m/z consistent with molecular formula C34H36O11 (DU = 17). The FTIR spectrum showed presence of OH (3508 cm−1), = CH (3373 cm−1), methyl group (2937 cm−1), carbonyl (1734 cm−1), acetate (1097 cm−1). The 1H, 13C and DEPT-135 showed the presence of 7 methyl, 2 methylene, 8 methine and 17 quaternary carbons. There are 4 protons signals at δ 6.58, 6.55, 6.10, 5.85, and 4 methoxy protons at δ 4.07, 4.07, 3.41, 3.37 along with 3 methyl protons at δ 1.18, 1.01, 1.06 indicated the presence of bioxanthracene scaffold. The literature search with this information suggested the structure of ES-242-3. The structure was further confirmed by comparing COSY, HSQC and HMBC correlations (Fig. 2c). ES-242-3 was earlier reported from soil fungus Penicillium sp., insect pathogenic fungus Cordyceps pseudomilitaris BCC 1620, and Verticillium sp. [20–22]. This compound is observed for the first time in the Talaromyces genus.
The anticancer activity of the isolated compounds was evaluated against three cancer cell lines MCF-7, HeLa, and CAL-27. 152G256α-1 showed the highest activity against oral cancer (CAL-27 cell line) with an IC50 value of 2.96 ± 0.17 µM than the remaining two compounds. ES-242-3 was the most active of all three molecules against cervical cancer (HeLa cell line) with IC50 4.46 ± 0.05 µM. The compounds were tested on normal human breast cell line MCF-10A and observed no toxicity up to 100 µM. The anti-cancer activities of isolated compounds are tabulated in Table 3.
Table 3.
In-vitro anti-cancer assay of compounds on MCF-7, Cal-27 and HeLa cell line (IC50 in μM)
| Sr. no. | Compound | MCF-7 | Cal-27 | HeLa |
|---|---|---|---|---|
| 1 | Peniazaphilin B | 51.98 ± 0.05 | 12.21 ± 0.3 | 10.38 ± 0.09 |
| 2 | 152G256α-1 | 24.01 ± 0.1 | 2.96 ± 0.17 | 5.09 ± 0.13 |
| 3 | ES-242-3 | 14.08 ± 0.2 | 9.05 ± 0.04 | 4.46 ± 0.05 |
| 4 | Doxorubicin | 1.24 ± 0.01 | 1.45 ± 0.01 | 1.16 ± 0.01 |
Molecular docking studies were carried out to unravel the interaction of the compounds with the selected cancer-associated protein targets. The prominently expressed receptors in breast, oral and cervical cancers were selected for docking studies namely ESR-1, WNT-7B, ALIX, TSG-101 and TFRC [23–26]. The binding energy for the molecule-ligand interaction, number of conventional H-bonds and hydrophobic interaction forming residues have been compiled in Tables 4, 5 and 6. 152G256α-1 showed interactions with ALIX and TSG-101 through 4 conventional H-bonds with amino acids Asp141, Asn 255, Lys 303, and Asp 299 (Fig. 3). The highest binding towards ESR1 protein was observed with ES-242-3 with the binding score of − 8.36 kcal/mol and forms 3 conventional H-bonds with amino acids Lys 530, Met 528, and Asp299 (Fig. 4). ES-242-3 binds with TFRC protein with a binding energy score of − 8.27 kcal/mol and forms 6 convention H-bonds with amino acids Lys240, Leu537, Gly703, Lys534, Trp702, and Asn270 (Fig. 5).
Table 4.
Molecular docking scores of isolated compounds against breast cancer-associated proteins
| Protein | Compound | Binding energy (kcal/mol) | Inhibition constant | Ligand efficiency | No. of conventional H-bonds | Hydrophobic interaction forming residues | The average distance of H Bonds (Å) |
|---|---|---|---|---|---|---|---|
| ESR1 (PDB ID: 3ERT) | Peniazaphilin B | − 6.5 | 17.29 | − 0.43 | 2(Lys449, Glu353) | Pro324 | 2.5 |
| 15G256α-1 | − 2.64 | 11.71 | − 0.66 | 3(Cys530, Glu419, Met522 |
Met528 Cys530 Val533 Trp350 Leu525 Met343 |
2.4 | |
| ES-242-3 | − 8.36 | 744.44 | − 0.19 | 3(Cys530, Met528, Asp351) | Leu525, Trp383, Cys530 | 2.5 | |
| WNT7B | Peniazaphilin B | − 5.81 | 55.12 | − 0.39 | 2(Trp82, Lys229) |
Phe79 Arg81 Ala301 |
2.7 |
| 15G256α-1 | − 6.74 | 11.39 | − 0.14 | 1(Ser124) |
His117 Ala121 Leu218 Lys220 Trp215 |
2.1 | |
| ES-242-3 | − 6.07 | 35.63 | − 0.13 | 3(Asn325, Gln323, Glu341) | Trp215, Cys324, Cys339 | 2.1 |
Table 5.
Molecular docking scores of compounds against oral cancer-associated proteins
| Protein | Compound | Binding energy (kcal/mol) | Inhibition constant | Ligand efficiency | No. of conventional H-bonds | Hydrophobic interaction forming residues | The average distance of H-Bonds (Å) |
|---|---|---|---|---|---|---|---|
| ALIX (PDB ID: 3C3R) | Peniazaphilin B | − 6.19 | 28.99 | − 0.41 | 4(Arg276, Leu326) | Pro331 | 2.6 |
| 15G256α-1 | − 7.81 | 1.9 | − 0.17 | 4(Asp141, Asn255, Lys303, Asp299) |
Leu140 Leu201 Phe4 Phe200 |
2.5 | |
| ES-242-3 | − 7.19 | 5.39 | − 0.16 | 4(Tyr294, Asn255, Lys303, Asp299) | Leu140, Leu201, Phe4, Phe200 | 2.2 | |
|
15G256a-1 TSG101 (PDB ID: 3OBQ) |
Peniazaphilin B | − 6.65 | 13.3 | − 0.44 | 1(Ser143) |
Phe142 Ile170 Pro139 Val161 Tyr68 |
2.0 |
| 15G256α-1 | − 6.94 | 8.2 | − 0.15 | 3(Thr58, Ser94) | Pro71, Met95, Lys36 | 2.9 | |
| ES-242-3 | − 6.35 | 22.01 | − 0.14 | - | Thr92 | - |
Table 6.
Molecular docking scores of isolated compounds against cervical cancer-associated proteins
| Protein | Compound | Binding energy (kcal/mol) | Inhibition constant | Ligand efficiency | No. of conventional H-bonds | Hydrophobic interaction forming residues | The average distance of H-Bonds (Å) |
|---|---|---|---|---|---|---|---|
| TFRC (PDB ID: 1DE4) | Peniazaphilin B | − 7.04 | 7.06 | − 0.47 | 4(Phe494, Lys531, Gln524, Asp525) | Met510 | 2.9 |
|
15G256α-1 (ID:) |
− 8.39 | 709.16 | − 0.18 | 5(Gln721, Asn273, Lys717) |
Lys240 Leu718 Gln721 Lys240 Leu274 Ala725 |
2.0 | |
| ES-242-3 | − 8.27 | 865.14 | − 0.18 | 6(Lys240, Leu537, Gly703, Lys534, Trp702, Asn270) |
Fig. 3.

2D interaction diagram of 152G256α-1 with Programmed cell death 6-interacting protein (ALIX)
Fig. 4.

2D interaction diagram of ES-242-3 with (ESR1) estrogen receptor
Fig. 5.

2D interaction diagram of Es-242-3 with Transferrin receptor protein (TFRC)
Further, drug likeliness and pharmacokinetic properties were estimated using numerous computational methods (Table 7). Although well-known marketed Natural Products drugs violate Lipinski’s rule of 5 in several aspects like molecular weight and hydrogen bond acceptor, this in-silico data can help to optimize the synthetic modification strategies and formulation aspects for these molecules. Peniazaphilin B follows Lipinski’s rule whereas, 152G256α-1 and ES-242-3 defy in terms of molecular weight, number of hydrogen bond acceptors, and topological surface area. Low to moderate blood–brain and Caco-2 permeability were observed for the isolated molecules. These compounds are CYP inhibitors namely CYP-2C19, CYP-2C9, and CYP-3A4 and hence should be cautiously used in combination therapy to avoid adverse reactions. It is important to note that these natural products showed weak plasma protein binding indicating a low toxicity index. Skin permeability of all compounds is well in the range making them suitable to be used in transdermal patches and other topical formulations as it will further avoid the first-pass metabolism like fentanyl transdermal patches for oral cancer [27].
Table 7.
The predicted pharmacokinetic and drug likeliness properties of the isolated compounds by computational tools
| Peniazaphilin B | 15G256α-1 | ES242-3 | |
|---|---|---|---|
| BBB permeability (2.0–0.1: moderate absorption; < 0.1: low absorption) |
0.650509 Cbrain/Cblood |
0.0522583 Cbrain/Cblood |
0.0160411 Cbrain/Cblood |
| Caco2 permeability (4–70 moderate permeability) | 20.2 nm/s | 15.4 nm/s | 22.8 nm/s |
| Skin permeability (log Kp; with Kp in cm/s; −9.7 < log Kp < −3.5) | − 6.25 cm/s | − 7.59 cm/s | − 7.38 cm/s |
| Subcellular localization | Mitochondria | Mitochondria | Mitochondria |
| CYP-Inhibition | |||
| CYP-2C19 | Inhibitor | Inhibitor | Inhibitor |
| CYP-2C9 | Inhibitor | Inhibitor | Inhibitor |
| CYP-2D6 | Non-inhibitor | Non-inhibitor | Non-inhibitor |
| CYP-2D6 | Non-inhibitor | Non-inhibitor | Non-inhibitor |
| CYP-3A4 | Inhibitor | Inhibitor | Inhibitor |
| Water solubility | 5160.46 mg/L | 409.16 mg/L | 0.01 mg/L |
| Plasma Protein Binding | 68.5% | 86.1% | 87.6% |
| Log P (≤ 5) | 1.63 | 2.54 | 4.69 |
| Hydrogen bond Donor (≤ 5) | 1 | 5 | 4 |
| Hydrogen Bond Acceptor (≤ 10) | 4 | 15 | 11 |
Conclusion
The report unveils three different types of molecules isolated from the ELF T. pinophilus cultured from the lichen Porina tetracerae. A hyphenated technique LC-UV-HRMS along with bioassay was applied for the selection of T. pinophilus out of all the screened 31 ELF species. These bioactive molecules are from azaphilone, macrocyclic polyester and bioxanthracene class of secondary metabolites. It was observed that molecule 15G256α-1 has shown potent oral cancer activity against CAL-27 cell-line with IC50 value 2.96 µM. The available drug for this treatment is cisplatin which showed an IC50 value of 5.3 µM against the same cell line. The macrocyclic ester along with azaphilones are produced by the polyketide synthase pathway. However, the precursor for the bioxanthracene molecule is not well established and hence warrants further investigation. Here, we report ES-242-3 from the bioxanthracene class for the first time from genus Talaromyces indicating the concealed potential of this genus to generate diverse chemical scaffolds. This study emboldens the scientists to explore the bioactive molecules associated with ELF.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We acknowledge the Department of Science and Technology for funding this project (DST Project No. DST/INT/SL/P-22/2016), Department of Pharmaceuticals, Ministry of Chemicals and Fertilizers, Government of India and National Institute of Pharmaceutical Education and Research (NIPER), Ahmedabad for providing the continuous support and facilities required for the work. CD analysis has been carried out at IIT Gandhinagar.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chaitrali Shevkar and Ashwini Armarkar have contributed equally to this work.
References
- 1.Vasan N, Baselga J, Hyman DM. A view on drug resistance in cancer. Nature. 2019;575:99–309. doi: 10.1038/s41586-019-1730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haider T, Pandey V, Banjare N, Gupta PN, Soni V. Drug resistance in cancer: mechanisms and tackling strategies. Pharmacol Rep. 2020;72:1125–1151. doi: 10.1007/s43440-020-00138-7. [DOI] [PubMed] [Google Scholar]
- 3.Gordaliza M. Natural products as leads to anticancer drugs. Clin Transl Oncol. 2007;9:767–776. doi: 10.1007/s12094-007-0138-9. [DOI] [PubMed] [Google Scholar]
- 4.Newman DJ, Cragg GM. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Nat Prod. 2020;83:770–803. doi: 10.1021/acs.jnatprod.9b01285. [DOI] [PubMed] [Google Scholar]
- 5.Schulz B, Draeger S, de Cruz TE, Rheinheimer J, Siems K, Loesgen S, Bitzer J, Schloerke O, Zeeck A, Kock I, Hussain H (2008) Screening strategies for obtaining novel, biologically active, fungal secondary metabolites from marine habitats. Bot Marine 51:219–234.
- 6.Müller K. Pharmaceutically relevant metabolites from lichens. Appl Microbial Biotechnol. 2001;56:9–16. doi: 10.1007/s002530100684. [DOI] [PubMed] [Google Scholar]
- 7.Singh BN, Upreti DK, Gupta VK, Dai XF, Jiang Y. Endolichenic fungi: a hidden reservoir of next generation biopharmaceuticals. Trends Biotechnol. 2017;35:808–813. doi: 10.1016/j.tibtech.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 8.Maduranga K, Attanayake RN, Santhirasegaram S, Weerakoon G, Paranagama PA. Molecular phylogeny and bioprospecting of Endolichenic Fungi (ELF) inhabiting in the lichens collected from a mangrove ecosystem in Sri Lanka. PLoS ONE. 2018 doi: 10.1371/journal.pone.0200711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blashfield RK, Aldenderfer MS. The literature on cluster analysis. Multivar Behavior Res. 1978;13:271–295. doi: 10.1207/s15327906mbr1303_2. [DOI] [PubMed] [Google Scholar]
- 10.Zhai MM, Li J, Jiang CX, Shi YP, Di DL, Crews P, Wu QX. The bioactive secondary metabolites from Talaromyces species. Nat Prod Bioprospect. 2016;6:1–24. doi: 10.1007/s13659-015-0081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakayama GR, Caton MC, Nova MP, Parandoosh Z. Assessment of the Alamar Blue assay for cellular growth and viability in vitro. J Immunol Methods. 1997;204:205–208. doi: 10.1016/s0022-1759(97)00043-4. [DOI] [PubMed] [Google Scholar]
- 12.UniProt Consortium The universal protein resource (UniProt) Nucleic Acids Res. 2007;36:D190–D195. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rezaei-Seresht H, Cheshomi H, Falanji F, Movahedi-Motlagh F, Hashemian M, Mireskandari E. Cytotoxic activity of caffeic acid and gallic acid against MCF-7 human breast cancer cells: an in silico and in vitro study. Avicenna J Phytomed. 2019;9:574. doi: 10.22038/AJP.2019.13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoda A, Tafaj M, Sallaku E. In silico structural, functional and phylogenetic analyses of cellulase from Ruminococcus albus. J Genet Eng Biotechnol. 2021;19:1–15. doi: 10.1186/s43141-021-00162-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huey R, Morris GM, Forli S. Using AutoDock 4 and AutoDock vina with AutoDockTools: a tutorial. The Scripps Research Institute Molecular Graphics Laboratory. 2012;105:92037. [Google Scholar]
- 16.Kadri A, Aouadi K. In vitro antimicrobial and α-glucosidase inhibitory potential of enantiopure cycloalkylglycine derivatives: insights into their in silico pharmacokinetic, druglikeness, and medicinal chemistry properties. J Appl Pharm Sci. 2020;10:107–115. doi: 10.7324/JAPS.2020.10614. [DOI] [Google Scholar]
- 17.Zhang D, Zhao J, Wang X, Zhao L, Liu H, Wei Y, You X, Cen S, Yu L. Peniazaphilin A, a new azaphilone derivative produced by Penicillium sp. CPCC 400786. J Antibiot. 2018;71:905–907. doi: 10.1038/s41429-018-0077-4. [DOI] [PubMed] [Google Scholar]
- 18.Dethoup T, Manoch L, Kijjoa A, Pinto M, Gales L, Damas AM, Silva AM, Eaton G, Herz W. Merodrimanes and other constituents from Talaromyces thailandiasis. J Nat Prod. 2007;70:1200–1202. doi: 10.1021/np0680578. [DOI] [PubMed] [Google Scholar]
- 19.He JW, Mu ZQ, Gao H, Chen GD, Zhao Q, Hu D, Sun JZ, Li XX, Li Y, Liu XZ, Yao XS. New polyesters from Talaromyces flavus. Tetrahed. 2014;70:4425–4430. doi: 10.1016/j.tet.2014.02.060. [DOI] [Google Scholar]
- 20.Gao H, Zhou L, Li D, Gu Q, Zhu TJ. New cytotoxic metabolites from the marine-derived fungus Penicillium sp. ZLN29. Helv Chim Acta. 2013;96:514–519. doi: 10.1002/hlca.201200596. [DOI] [Google Scholar]
- 21.Isaka M, Kongsaeree P, Thebtaranonth Y. Bioxanthracenes from the insect pathogenic fungus Cordyceps pseudomilitaris BCC 1620 II. Structure elucidation. J Antibiot. 2001;54:36–43. doi: 10.7164/antibiotics.54.36. [DOI] [PubMed] [Google Scholar]
- 22.Toki S, Ando K, Kawamoto I, Sano H, Yoshida M, Matsuda Y. ES-242-2,-3,-4,-5,-6,-7, and-8, novel bioxanthracenes produced by Verticillium sp., which act on the N-methyl-D-aspartate receptor. J Antibiot. 1992;45:1047–1054. doi: 10.7164/antibiotics.45.1047. [DOI] [PubMed] [Google Scholar]
- 23.Holst F, Stahl PR, Ruiz C, Hellwinkel O, Jehan Z, Wendland M, Lebeau A, Terracciano L, Al-Kuraya K, Jänicke F, Sauter G. Estrogen receptor alpha (ESR1) gene amplification is frequent in breast cancer. Nat Genet. 2007;39:655–660. doi: 10.1038/ng2006. [DOI] [PubMed] [Google Scholar]
- 24.Nakamichi E, Sakakura H, Mii S, Yamamoto N, Hibi H, Asai M, Takahashi M. Detection of serum/salivary exosomal Alix in patients with oral squamous cell carcinoma. Oral Dis. 2021;27:439–447. doi: 10.1111/odi.13565. [DOI] [PubMed] [Google Scholar]
- 25.Leung LL, Riaz MK, Qu X, Chan J, Meehan K (2021) Profiling of extracellular vesicles in oral cancer, from transcriptomics to proteomics. In: Seminars in cancer biology. Academic Press. 10.1016/j.semcancer.2021.01.002 [DOI] [PubMed]
- 26.Xu X, Liu T, Wu J, Wang Y, Hong Y, Zhou H. Transferrin receptor-involved HIF-1 signalling pathway in cervical cancer. Cancer Gene Ther. 2019;26:356–365. doi: 10.1038/s41417-019-0078-x. [DOI] [PubMed] [Google Scholar]
- 27.Ahn JS, Lin J, Ogawa S, Yuan C, O’Brien T, Le BH, Bothwell AM, Moon H, Hadjiat Y, Ganapathi A. Transdermal buprenorphine and fentanyl patches in cancer pain: a network systematic review. J Pain Res. 2017;10:1963. doi: 10.2147/JPR.S140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.