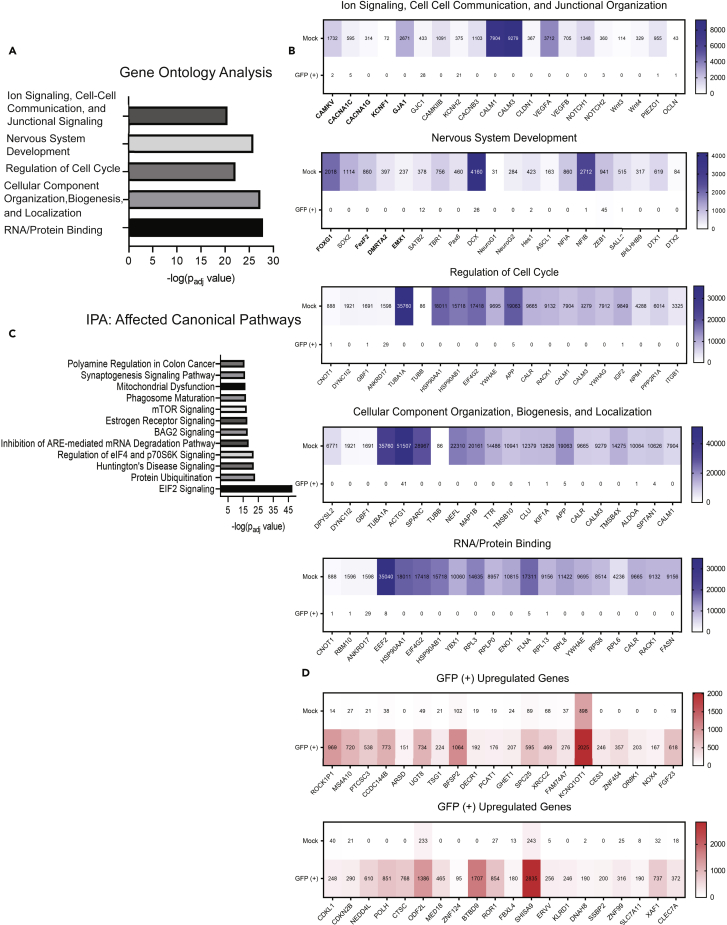
Figure 4.
Pathway and ontology analysis conducted on GFP (+) vs. Mock and heatmaps displaying up- and downregulated genes
(A) Gene Ontology analysis of the top 3,000 differentially expressed genes found from DESEQ2 analysis in R of GFP (+) vs. Mock using cutoffs of fold change ± 3 and adj p value < 0.01. Terms listed were the five most significant across categories molecular function, biological process, and cellular components (for complete list see Table S5).
(B) Heatmaps showing 20 representative genes within the ontology classifications from (A); the values in the heatmap are the average raw read count number across GFP (+) or Mock samples.
(C) Most significantly impacted canonical pathways as determined by Ingenuity Pathway Analysis using the same gene list as in (A, see Table S6 for complete list).
(D) Heatmaps containing 20 of the most significantly upregulated genes in GFP (+) and GFP (Low) groups vs. Mock determined by adj p value and fold change. For ontology and IPA, significance is displayed as -log (adj p-value) using cutoffs stated above. Bolded genes within the heatmaps identify those used in subsequent Shield-1 experiments. Also see Figures S3, S4A–S4C, and S5 along with Tables S5 and S6.