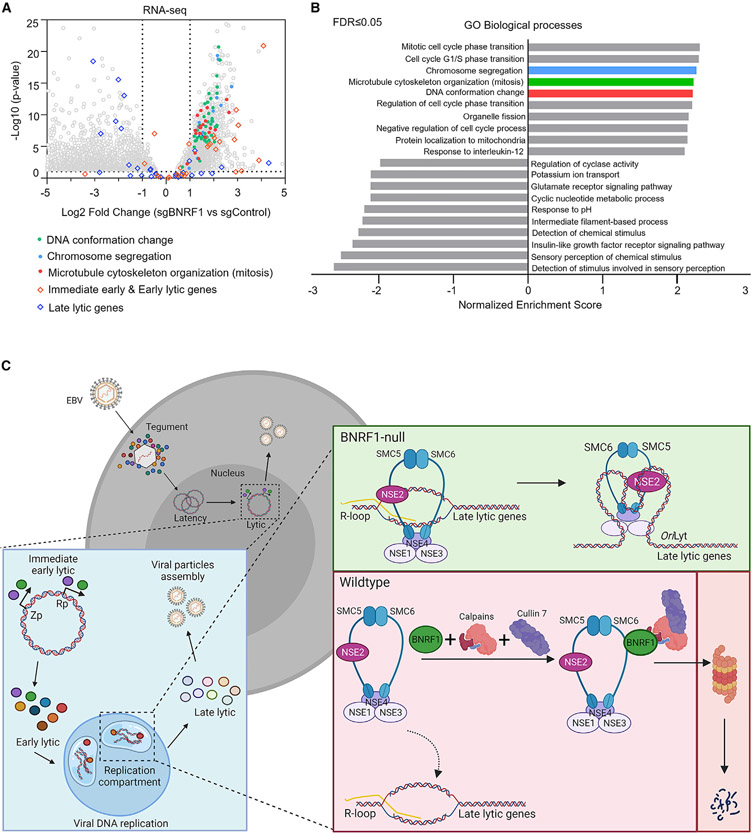
Figure 7. BNRF1 KO effects on lytic cycle host and viral gene expression.
(A) RNA-seq volcano plot analysis of host mRNA abundance in P3HR-1 ZHT/RHT cells that expressed BNRF1 versus control sgRNAs and that were induced by 4-HT/NaB for 24 h, as in Figure 4A. Circles represent individual host mRNA values. Transcripts encoding proteins involved in DNA conformation change, chromosome segregation, and mitosis are highlighted.
(B) Gene Ontology (GO) biological process enrichment analysis of host mRNAs differentially expressed in P3HR-1 ZHT/RHT cells with BNRF1 versus control sgRNAs and induced by 4-HT/NaB for 24 h, as in Figure 4A.
(C) Schematic of BNRF1 evasion of SMC5/6 complexes in lytic replication. BNRF1 produced upon late lytic cycle progression associates with calpain and Cul7 to target the SMC5/6 cohesin complex for ubiquitin-proteasome pathway degradation. In the absence of BNRF1, the SMC5/6 complex associates with EBV genomic R-loops, including at the oriLyt and perhaps additional non-B-form DNA structures formed in lytic replication to prevent sustained late gene expression, RC formation, EBV lytic genome encapsidation, and infectious virion assembly.