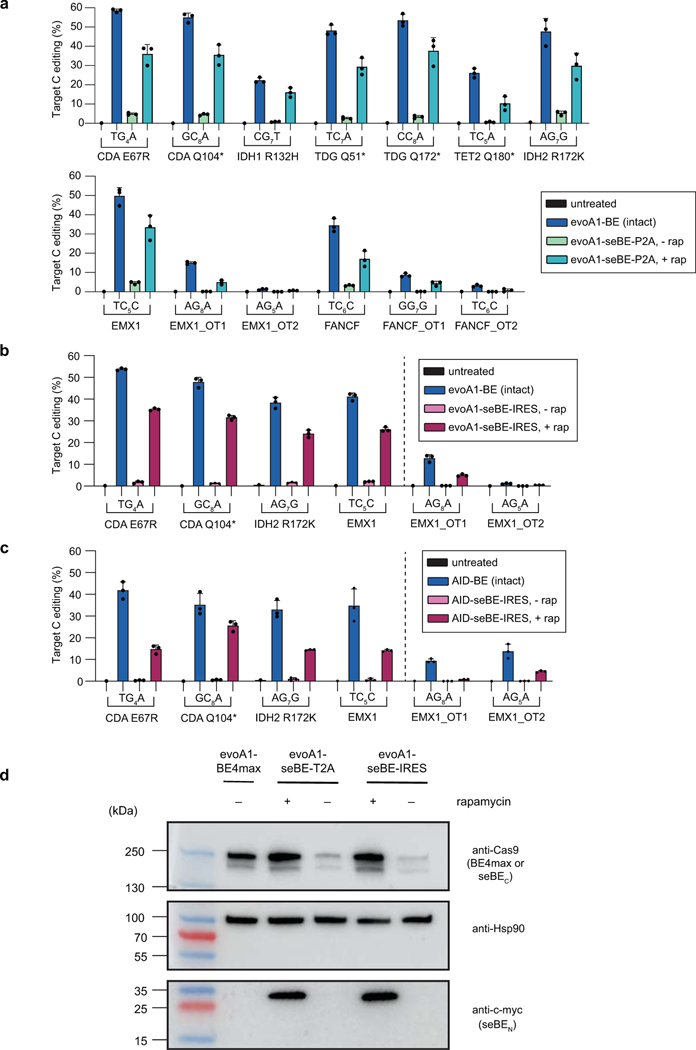
Extended Data Fig. 5. Base editing efficiency in human cells at diverse loci and off-target sites.
HEK293T cells were untreated or transfected with intact BE4max or seBE constructs in the absence or presence of rapamycin. C or G describes whether the coding (C) or non-coding (G) strand cytosine is targeted, respectively, with the subscript denoting the position of the sgRNA targeted by the base editor. Shown are experiments with control constructs and (a) evoA1-seBE-T2A (corresponding to Fig. 3b and Fig. 4b), (b) evoA1-seBE-IRES (corresponding to Fig. 3c and Fig. 4b), and (c) AID’-seBE-IRES (corresponding to Fig. 3c and Fig. 4b). Bars indicate mean and error bars indicate standard deviations of n = 3 biological replicates. Complete editing footprints for each locus and replicate in (a-c) are provided in Supplementary Figs. 1-3. (d) Western blot of cells transfected with evoA1-based constructs in the absence and presence of rapamycin. The Cas9 antibody probes the intact editor and the seBEC fragment (which are of similar size); Hsp90 antibody serves as a loading control; c-myc antibody probe the N-terminal tag of the seBEN fragment. Representative experiment was repeated independently two times and the results were reproducible.