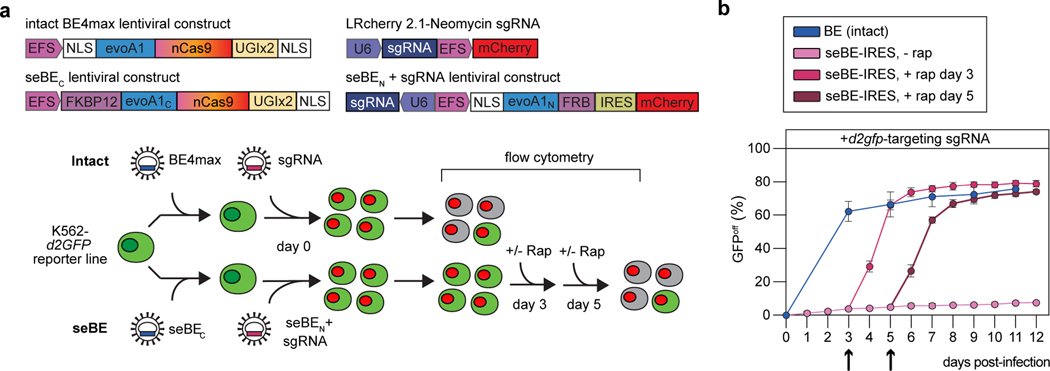
Figure 5. seBEs permit temporal control over base editing.
(a) Top—shown are the lentiviral constructs for introducing the intact BE4max or the seBE fragments along with mCherry expressing sgRNA constructs. Bottom—editing efficiency was evaluated in a K562 cell line containing a single copy of integrated, constitutively expressed d2gfp. Editing of the d2gfp locus results in loss of GFP fluorescence which can be tracked by flow cytometry either in the presence or absence of added rapamycin. (b) Quantification of GFPoff cells by flow cytometry for cells with intact BE4max or seBE-IRES with no rapamycin or rapamycin (25 nM) added at either day 3 or day 5 (marked with arrow) and then maintained continuously. Mean and standard deviation are noted, with individual data points shown (n = 3).