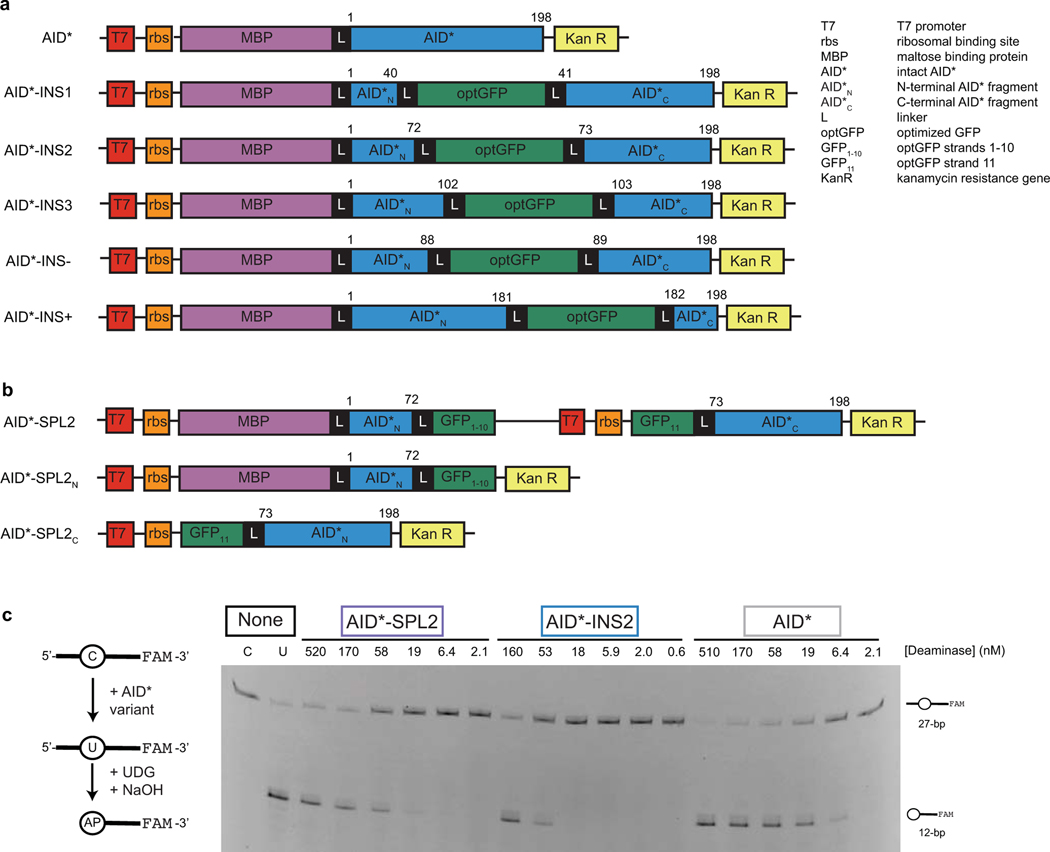
Extended Data Fig. 1. Intact, inserted, and split DNA deaminase constructs with AID*.
(a) Construct schematics for AID* and AID*-INS variants (analogous to those in Fig. 1b) that were used to determine the impact of optGFP insertion in E. coli. Numbers above the constructs represent the amino acid sequence of the deaminase itself. (b) Construct schematics for co-expression or individual expression of AID*-SPL2N and AID*-SPL2C fragments in E. coli. (c) Left—an in vitro assay to measure deaminase activity on a 3’-fluorescein (FAM) labeled oligonucleotide substrate. UDG, uracil DNA glycosylase. Right—a representative denaturing gel (100 nM DNA, variable enzyme concentration) is shown, along with substrate and product controls (C and U, respectively). Analysis of product formation with three independent replicates was used to define the dose-response curve provided in Fig. 1d.