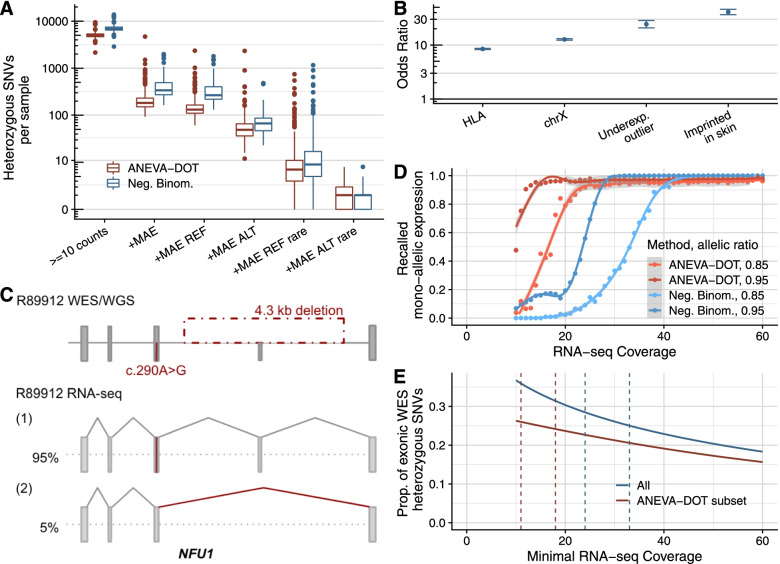
Fig. 5.
Mono-allelic expression. A Distribution of heterozygous SNVs per sample for successive filtering steps from left to right: Heterozygous SNVs detected by WES with an RNA-seq coverage of at least 10 reads, where MAE is detected, where MAE of the reference is detected, where MAE of the alternative is detected, and subsetted for rare variants. MAE expression detected using ANEVA-DOT and a negative binomial test (Methods). B Odds ratio of MAE in genes with common variants only and with at least one rare variant across different gene categories. Results shown for the negative binomial test only. Error bars represent 95% confidence intervals of pairwise logistic regressions. C Schematic depiction of the disease-causing 4.3 kb deletion and the c.290A>G SNV in NFU1, and their consequence on the RNA level in sample R89912. The percentage of each transcript isoform is shown next to it. Figure not shown at genomic scale. D Fraction of recalled MAE events (FDR < 0.05 on each method) with simulated allelic ratios of 0.85 and 0.95 as function of RNA-seq coverage. E Proportion of exonic heterozygous WES SNVs detected in all genes as a function of minimal RNA-seq coverage. ANEVA-DOT is able to detect only a subset of SNVs. Vertical lines correspond to RNA-seq coverage needed to recall 90% of simulated allelic ratios of 0.85 and 0.95 as inferred from panel D. REF: reference, ALT: alternative, rare: minor allele frequency < 0.1%