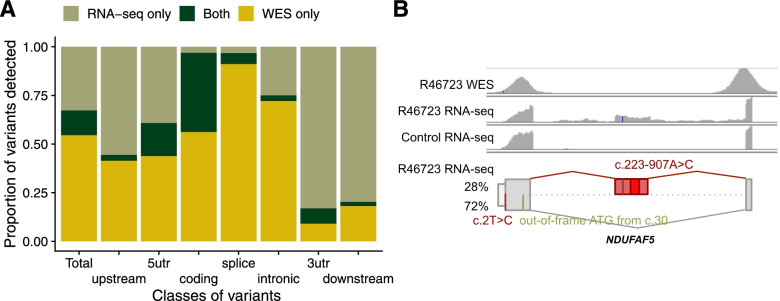
Fig. 6.
RNA-seq variant calling. A Median across samples of the proportion of variants called only by WES, only by RNA-seq, and by both technologies, in total and stratified by variant classes. Of note, over 50% of variants in coding regions are called only by WES, probably because of RNA-seq limitations including that not all the genes are expressed in fibroblasts, the uneven read coverage along the transcript, and because the expression level of variant-carrying alleles must be high enough to yield sufficient RNA-seq read coverage. B WES (row 1) and RNA-seq (row 2) coverage of the affected sample (R46723) and a representative control (row 3) using IGV. The created exon and a variant are seen in the affected RNA profile, but not covered in the corresponding WES and not present in the control. Bottom row: schematic depiction of the NM_024120.4 c.2T>C and c.223-907A>C variants and their consequence on the RNA level with an out-of-frame ATG (in green), and a cryptic exon with the PTC (bright red rectangle) on NDUFAF5. The percentage of detected transcript isoform is shown next to it. Figure not shown at genomic scale