FIGURE 4.
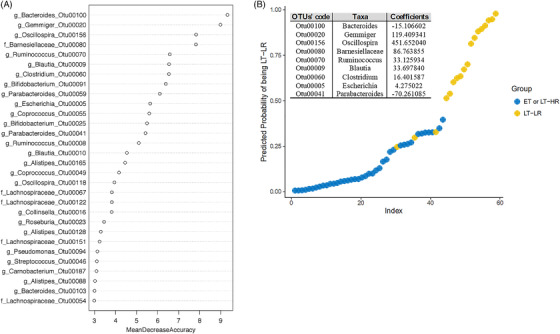
Calculations for an abundance‐based operational taxonomic units (OTUs) signature to predict immune progression of human immunodeficiency virus (HIV)‐infected subjects from their gut mucosal microbiota. Relative abundances from rarefied counts were used as input to 1000 random forests, and out‐of‐bag error was computed using the randomForest package in R. Initial analysis was performed with samples of the three treated HIV‐groups separated, but a classification error of 73% with LT‐HR samples, as many of them were assimilated as ET, and an out‐of‐bag error of 35% uncovered the great overlap existing between ET and LT‐HR groups. Once grouped together for further analyses, the out‐of‐bag error decreased to 12%. Given the multi‐collinearity of some OTUs, Lasso (L1‐norm) regularization was performed using R glmnet package, selecting the lambda through k‐3 cross‐validation, which minimizes the error in order to remove less relevant and multi‐collinear features (OTUs) before performing the logistic regression analysis. ROC curves were computed using InformationValue package in R. (A) Random forest (RF) analysis showing the best 30 OTUs to classify samples in either ET/LT‐HR or LT‐LR groups, showing their respective mean decrease accuracies (MDA). (B) Multivariable logistic regression model built from the 14 best predictors in RF (MDA > 5) yielding a signature of nine OTUs, whose log‐odds coefficients are shown in the table, that discriminate samples belonging to ET/LT‐HR or LT‐LR groups with a minimum misclassification error of 5% and area under curve (AUC) of .97 in an ROC analysis. ET, early‐treated; LT‐HR, late‐treated high recovery; LT‐LR, late‐treated low recovery; f, family; g, genus
