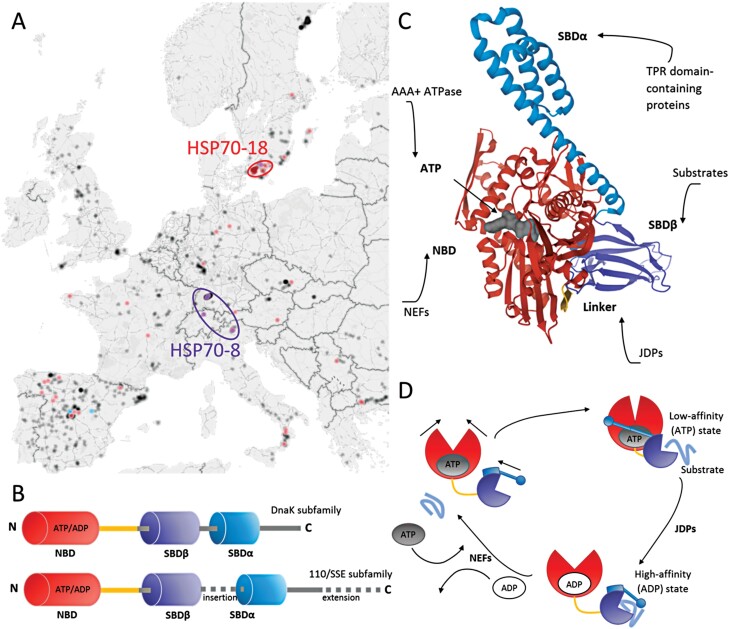
Fig. 1.
HSP70 sequence conservation, domain structure, and chaperone activity. (A) The frequency of mutations in HSP70-8 and HSP70-18 coincides with geographical location. The geographical distribution of all Arabidopsis accessions collected in the 1001 genome database (black) overlaid with the occurrence of high-impact mutations in HSP70-8 (purple), HSP70-13 (blue, no significant clustering), and HSP70-18 (red). Data retrieved from the 1001 genome database (Weigel and Mott, 2009) and visualized using a Microsoft Excel 3D map. (B) Domain organization of DnaK and 110/SSE subfamilies (Lin et al., 2001; Sarkar et al., 2013). (C) Structure of HSP70 with bound ATP. Modeled using the crystal structure of bacterial DnaK (PDB ID: 4B9Q; Kityk et al., 2012). Red, nucleotide-binding domain (NBD); blue, substrate-binding domain SBDα; violet, substrate-binding domain SBDβ; gray, ATP; yellow, linker. Sites of protein–protein interactions with HSP70 co-chaperones are indicated, including tetratricopeptide repeat (TPR) proteins, J-domain proteins (JDP), and nucleotide exchange factors (NEF). (D) Chaperone cycle of HSP70s. The interaction with JDPs stimulates ATP hydrolysis and the transition to the ADP-bound state with a high affinity for protein substrates. NEFs facilitate the HSP70 conversion back to the low-affinity state, the dissociation of ADP and rebinding of ATP, and the release of the protein substrate (Rosenzweig et al., 2019).