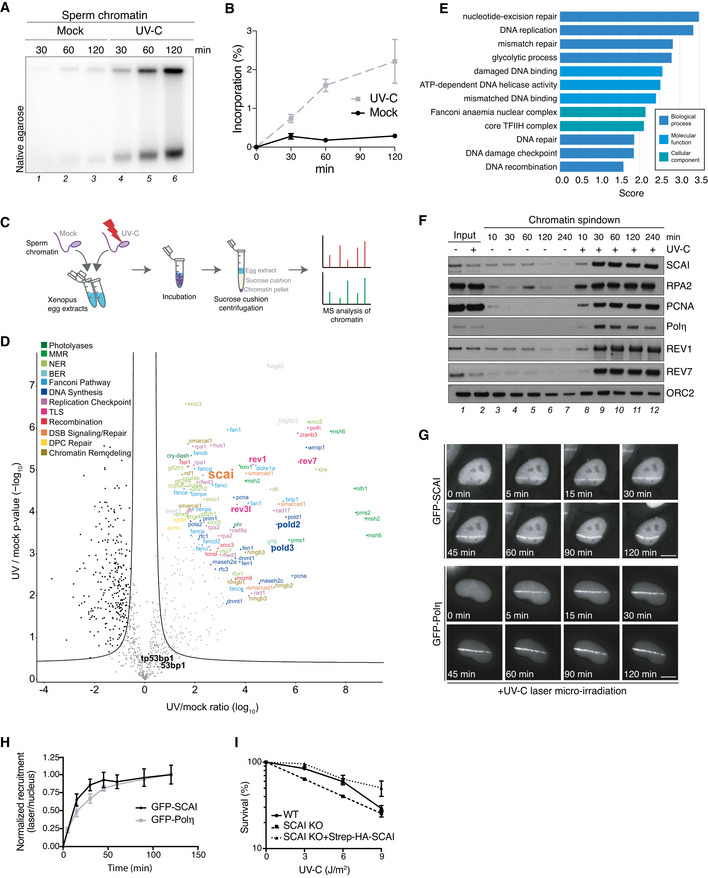
Sperm chromatin was left untreated or exposed to UV‐C (2,000 J/m2), incubated in HSS/NPE mix in the presence of [α‐32P]dATP and nucleotide incorporation into chromatin was analyzed at the indicated time points via native agarose gel electrophoresis.
Quantification of data in (A) (mean ± SEM; n = 3 independent experiments).
Schematic outline of CHROMASS workflow to analyze protein recruitment to UV‐damaged chromatin. Sperm chromatin left untreated or irradiated with UV‐C (2,000 J/m2) was incubated in Xenopus egg extract for 30 min, isolated by sucrose cushion centrifugation, and analyzed by label‐free mass spectrometry (MS).
Protein recruitment to UV‐damaged chromatin compared to an undamaged control. Volcano plot shows enrichment of individual proteins (UV/mock ratio) plotted against the P‐value (n = 4 independent experiments; FDR < 0.05, s0
= 0.5).
Term enrichment analysis showing GO terms corresponding to proteins significantly recruited to UV‐C‐damaged chromatin (Dataset
EV5). All displayed terms were significant with
P < 0.02, as determined through Fisher exact testing with Benjamini–Hochberg correction.
Sperm chromatin left untreated or irradiated with UV‐C (2,000 J/m2) was isolated at the indicated time points and analyzed by immunoblotting.
Representative images of GFP‐SCAI and GFP‐Polη recruitment to UV‐C laser micro‐irradiation in U2OS cells at indicated time points. Scale bar, 10 µm.
U2OS cells stably expressing GFP‐SCAI or transiently transfected with GFP‐Polη were subjected to UV‐C laser micro‐irradiation. At indicated time points, GFP fluorescence intensities at damage sites over the nuclear background were quantified. Recruitment was normalized to the time point of maximal recruitment (mean ± SEM; n = 3 independent experiments; at least 30 cells analyzed per condition).
Clonogenic survival of U2OS cells subjected to indicated doses of UV‐C (mean ± SEM; n = 3 independent experiments).