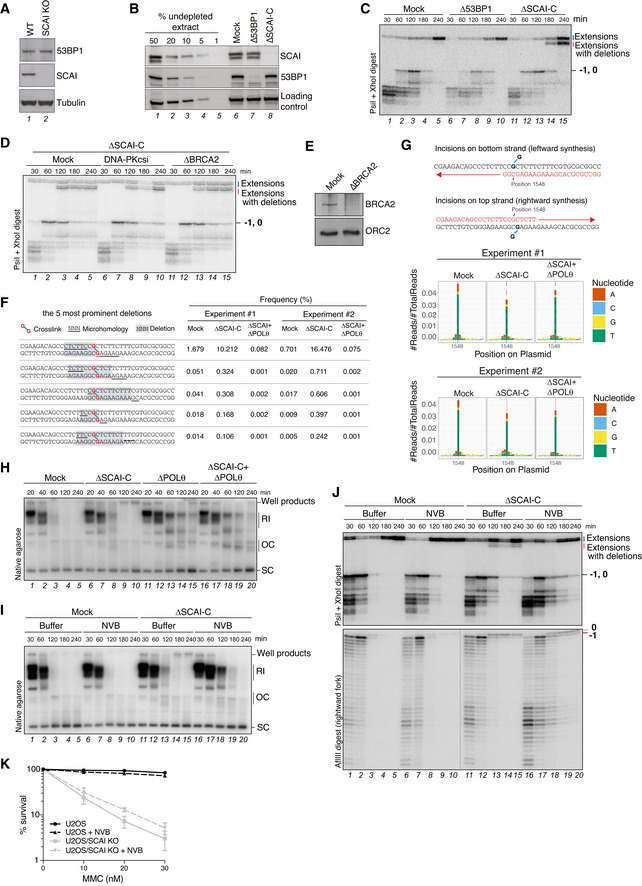
Whole cell extracts of U2OS and U2OS/SCAI KO cells were analyzed by immunoblotting with indicated antibodies.
Mock‐, 53BP1‐, or SCAI‐C‐depleted egg extracts were analyzed by immunoblotting with indicated antibodies.
pICLpt was replicated in extracts immunodepleted in mock‐, 53BP1‐, or SCAI‐C‐depleted egg extracts digested with PsiI+XhoI and resolved on a denaturing polyacrylamide gel.
pICLpt was replicated in SCAI‐C‐depleted extracts that were also depleted of BRCA2, or supplemented with DNA‐PKcs inhibitor (NU7441; 100 µM) where indicated. Reaction samples were digested with PsiI+XhoI and analyzed on a denaturing polyacrylamide gel.
Mock‐ or BRCA2‐depleted egg extracts were analyzed by immunoblotting with indicated antibodies.
Most frequent deletion products in sequencing data in Fig
5A. In red, crosslinked guanines; highlighted in grey, deletions; underlines, microhomology regions.
Schematic representation of DNA synthesis occurring across the adducted base following DNA incisions on either the top or bottom strand. The denoted nucleotide position 1,548 corresponds to the insertion 0 position when incisions occur in the bottom strand or to the −1 position when incisions occur in the top strand. Sequencing products of correct length from Fig
5A were analyzed for their nucleotide misincorporation distribution (bottom graphs). Misincorporation is based on the bottom strand read.
Samples in Fig
5C were analyzed by native agarose gel electrophoresis.
pICLpt was replicated in mock‐ or SCAI‐depleted extracts treated or not with Novobiocin (NVB; 150 µM) in the presence of [α‐32P]dATP, and reactions were analyzed by native agarose gel electrophoresis. RI, replication intermediates; OC, open circular; SC, supercoiled.
Samples in (I) were digested with PsiI+XhoI or AflIII and analyzed on a denaturing polyacrylamide gel.
Clonogenic survival of U2OS and U2OS/SCAI KO cells treated or not with Novobiocin (NVB; 50 µM) and subjected to various doses of MMC for 24 h (mean ± SEM; n = 3 independent experiments).