Figure 3. Cryo‐EM maps of S‐2P spike in complex with Sb#15 and Sb#68.
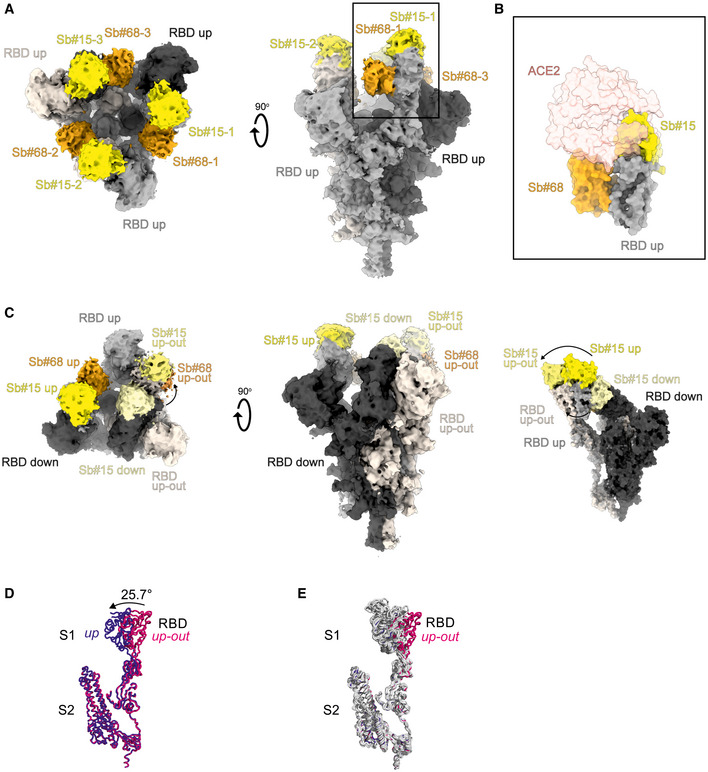
- Cryo‐EM map of S‐2P with both Sb#15 and Sb#68 bound to each RBD adopting a symmetrical 3up conformation.
- Close‐up view showing that ACE2 binding to RBD (PDB ID: 6M0J) is blocked by bound Sb#15 and by a steric clash with Sb#68.
- Cryo‐EM map of S‐2P with the three RBDs adopting an asymmetrical 1up/1up‐out/1down conformation. Sb#15 is bound to all three RBDs, while Sb#68 is only bound to the up and up‐out RBD. Final maps blurred to a B factor of −30 Å were used for better clarity of the less resolved RBDs and sybodies. Spike protein is shown in shades of gray, Sb#15 in yellow and Sb#68 in orange.
- Alignment of structural models for the up (blue) and up‐out (magenta) spike conformations. For clarity, only monomers are shown.
- The up‐out RBD conformation is unique among reported spike structures. Superposition of the aligned models (D) with 15 published structures of spike monomers showing up‐RBDs (gray). PDB identifiers of aligned structures: 6VSB, 6VYB, 6XKL, 6ZGG, 6ZXN, 7A29, 7B18, 7CHH, 7CWT, 7DX9, 7JWB, 7LWW, 7M6F, 7N0H, and 7N1V.
