Figure 5. The bispecific construct GS4 mitigates emergence of novel escape mutants.
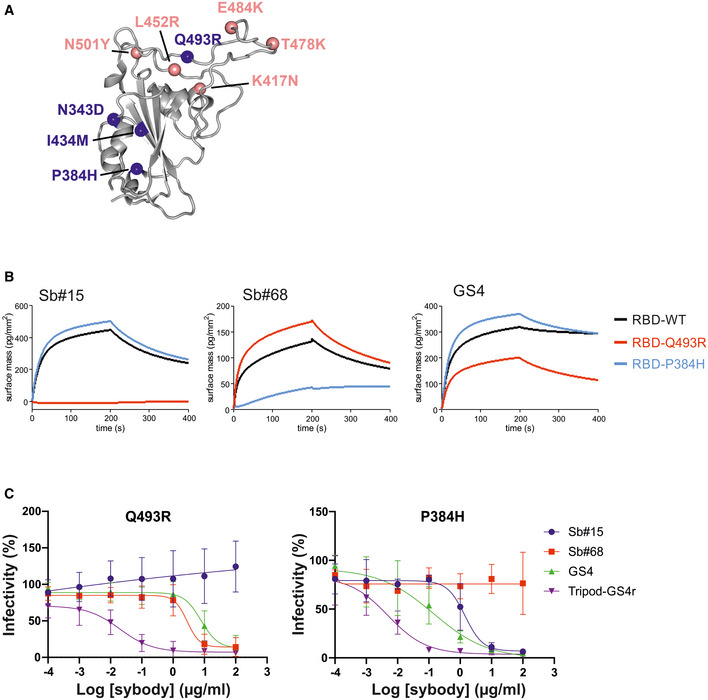
- Structural context of RBD escape mutants resulting from adaptation experiments in the presence of either Sb#15 or Sb#68 alone (blue spheres). Prominent globally circulating mutations are shown as salmon spheres.
- GCI‐based kinetic analysis of the purified RBD bearing either no mutation (WT), or identified escape mutations Q493R or P384H. Left, middle, and right plots correspond to immobilized Sb#15, Sb#68, or GS4, respectively.
- Neutralization assay using VSVΔG pseudotyped with Spike protein containing the Q493R or P384H mutation, respectively. Relative infectivity in response to increasing binder concentrations was determined. Error bars correspond to standard deviations of three biological replicates.
